VANTED: a system for advanced data analysis and visualization in the context of biological networks
- PMID: 16519817
- PMCID: PMC1413562
- DOI: 10.1186/1471-2105-7-109
VANTED: a system for advanced data analysis and visualization in the context of biological networks
Abstract
Background: Recent advances with high-throughput methods in life-science research have increased the need for automatized data analysis and visual exploration techniques. Sophisticated bioinformatics tools are essential to deduct biologically meaningful interpretations from the large amount of experimental data, and help to understand biological processes.
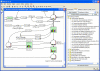
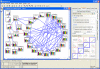
Results: We present VANTED, a tool for the visualization and analysis of networks with related experimental data. Data from large-scale biochemical experiments is uploaded into the software via a Microsoft Excel-based form. Then it can be mapped on a network that is either drawn with the tool itself, downloaded from the KEGG Pathway database, or imported using standard network exchange formats. Transcript, enzyme, and metabolite data can be presented in the context of their underlying networks, e. g. metabolic pathways or classification hierarchies. Visualization and navigation methods support the visual exploration of the data-enriched networks. Statistical methods allow analysis and comparison of multiple data sets such as different developmental stages or genetically different lines. Correlation networks can be automatically generated from the data and substances can be clustered according to similar behavior over time. As examples, metabolite profiling and enzyme activity data sets have been visualized in different metabolic maps, correlation networks have been generated and similar time patterns detected. Some relationships between different metabolites were discovered which are in close accordance with the literature.
Conclusion: VANTED greatly helps researchers in the analysis and interpretation of biochemical data, and thus is a useful tool for modern biological research. VANTED as a Java Web Start Application including a user guide and example data sets is available free of charge at http://vanted.ipk-gatersleben.de.
Figures


References
-
- Celis JE, Kruhoffer M, Gromova I, Frederiksen C, Ostergaard M, Thykjaer T, Gromov P, Yu JS, Palsdottir H, Magnusson N, Orntoft TF. Gene expression profiling: monitoring transcription and translation products using DNA microarrays and proteomics. FEBS Letters. 2000;480:2–16. doi: 10.1016/S0014-5793(00)01771-3. - DOI - PubMed
-
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell. 2004;16:3304–3325. doi: 10.1105/tpc.104.025973. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

