Hyaluronidases: their genomics, structures, and mechanisms of action
- PMID: 16522010
- PMCID: PMC2547145
- DOI: 10.1021/cr050247k
Hyaluronidases: their genomics, structures, and mechanisms of action
Figures

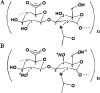
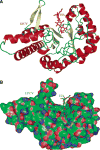
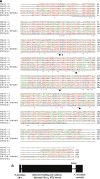
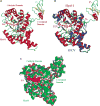
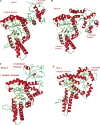
References
-
- Rigden DJ, Jedrzejas MJ. J Biol Chem. 2003;278:50596–606. - PubMed
-
- Jedrzejas MJ. Crit Rev Biochem Mol Biol. 2000;35:221–51. - PubMed
-
- Jedrzejas MJ. Front Biosci. 2004;9:891–914. - PubMed
-
- Jedrzejas MJ. In: Hyaluronan 2003. Hascall VC, Balazs EA, editors. Matrix Bilogy Institute; Edgewater, NJ: 2004. www.matrixbiologyinstitute.org/ha03.
-
- Jedrzejas MJ. In: Recent Research Developments in Biophysics and Biochemistry. Pandali SG, editor. Vol. 2 Research Signpost; Trivandrum, India: 2002.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

