Crystal structure of 3-hydroxyanthranilic acid 3,4-dioxygenase from Saccharomyces cerevisiae: a special subgroup of the type III extradiol dioxygenases
- PMID: 16522801
- PMCID: PMC2242480
- DOI: 10.1110/ps.051967906
Crystal structure of 3-hydroxyanthranilic acid 3,4-dioxygenase from Saccharomyces cerevisiae: a special subgroup of the type III extradiol dioxygenases
Abstract
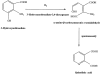
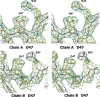
3-Hydroxyanthranilic acid 3,4-dioxygenase (3HAO) is a non-heme ferrous extradiol dioxygenase in the kynurenine pathway from tryptophan. It catalyzes the conversion of 3-hydroxyanthranilate (HAA) to quinolinic acid (QUIN), an endogenous neurotoxin, via the activation of N-methyl-D-aspartate (NMDA) receptors and the precursor of NAD(+) biosynthesis. The crystal structure of 3HAO from S. cerevisiae at 2.4 A resolution shows it to be a member of the functionally diverse cupin superfamily. The structure represents the first eukaryotic 3HAO to be resolved. The enzyme forms homodimers, with two nickel binding sites per molecule. One of the bound nickel atoms occupies the proposed ferrous-coordinated active site, which is located in a conserved double-strand beta-helix domain. Examination of the structure reveals the participation of a series of residues in catalysis different from other extradiol dioxygenases. Together with two iron-binding residues (His49 and Glu55), Asp120, Asn51, Glu111, and Arg114 form a hydrogen-bonding network; this hydrogen-bond network is key to the catalysis of 3HAO. Residues Arg101, Gln59, and the substrate-binding hydrophobic pocket are crucial for substrate specificity. Structure comparison with 3HAO from Ralstonia metallidurans reveals similarities at the active site and suggests the same catalytic mechanism in prokaryotic and eukaryotic 3HAO. Based on sequence comparison, we suggest that bicupin of human 3HAO is the first example of evolution from a monocupin dimer to bicupin monomer in the diverse cupin superfamilies. Based on the model of the substrate HAA at the active site of Y3HAO, we propose a mechanism of catalysis for 3HAO.
Figures

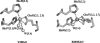




References
-
- Anand R., Dorrestein P.C., Kinsland C., Begley T.P., Ealick S.E. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution Biochemistry 41: 7659–7669. - PubMed
-
- Bugg T.D.H. and Lin G. 2001. Solving the riddle of the intradiol and extradiol catechol dioxygenases: How do enzymes control hydroperoxide rearrangements? Chem. Commun. 2001: 941–952.
-
- Bugg T.D., Sanvoisin J., Spence E.L. 1997. Exploring the catalytic mechanism of the extradiol catechol dioxygenases Biochem. Soc. Trans. 25: 81–85. - PubMed
-
- Calderone V., Trabucco M., Menin V., Negro A. 2002. Cloning of human 3-hydroxyanthranilic acid dioxygenase in Escherichia coli: Characterisation of the purified enzyme and its in vitro inhibition by Zn2+ Biochim. Biophys. Acta 1596: 283–292. - PubMed
-
- Cleasby A., Wonacott A., Skarzynski T., Hubbard R.E., Davies G.J., Proudfoot A.E., Bernard A.R., Payton M.A., Wells T.N. 1996. The x-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 angstrom resolution Nat. Struct. Biol. 3: 470–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

