Concentration and desalting of peptide and protein samples with a newly developed C18 membrane in a microspin column format
- PMID: 16522865
- PMCID: PMC2291762
Concentration and desalting of peptide and protein samples with a newly developed C18 membrane in a microspin column format
Abstract
Protein identification plays an important role in today's academic and industrial proteomic research. Commonly used methods for the separation of proteins from complex samples include liquid chromatography (e.g., ion exchange, reversed-phase, hydrophobic interaction), or types of gel electrophoresis (e.g., 1d and 2d PAGE). Relevant proteins separated in the latter way are often cut out, cleaved with trypsin "in gel," and the resulting peptide mixtures combined with matrix and spotted onto a target plate for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF-ms) analysis. Subsequently, proteins can be identified by comparison of the resulting peptide mass fingerprints against different databases.(1) since the success of protein identification can be enhanced by the desalting and concentration of the samples, an innovative C18-membrane was incorporated into a microspin column (Vivapure C18 micro spin column, Vivascience AG, Hannover, Germany) to analyze its performance for sample preparation prior to MALDI-ToF-ms. Rapid concentration of single or multiple 200-microl volumes through an available membrane only 2 mm in diameter allowed for analysis of very dilute samples. We observed the successful and rapid desalting of urea-containing protein samples at 100 fmol/mul up to a mass of approximately 70 KDA and the concentration of digest peptides from a solution of 1 fmol/microl using C18-membrane technology.
Figures
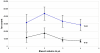
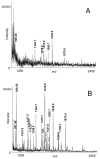
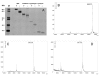
References
-
- Pappin DJ. Peptide mass fingerprinting using MALDI-ToF mass spectrometry. Methods Mol Biol 1997;4:165–173. - PubMed
-
- Nemeth-Cawley JF, Tangarone BS, Rouse JC. “Top Down” characterization is a complementary technique to peptide sequencing for identifying protein species in complex mixtures. J Proteome Res 2003;2:495–505. - PubMed
-
- Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics 2004;4:3665–3685. - PubMed
-
- Stone KL, Williams KR. Enzymatic digestion of Proteins and HPLC peptide isolation. In Matsudaira P (ed): A Practical Guide to Protein and Peptide Purification for Microsequencing, 2nd ed., San Diego: Academic Press, 1993:43–69.
-
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999; 20:3551–3567. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
