The origins and evolution of functional modules: lessons from protein complexes
- PMID: 16524839
- PMCID: PMC1609335
- DOI: 10.1098/rstb.2005.1807
The origins and evolution of functional modules: lessons from protein complexes
Abstract
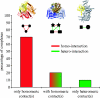
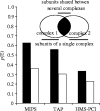
Modularity is an attribute of a system that can be decomposed into a set of cohesive entities that are loosely coupled. Many cellular networks can be decomposed into functional modules-each functionally separable from the other modules. The protein complexes in physical protein interaction networks are a good example of this, and here we focus on their origins and evolution. We investigate the emergence of protein complexes and physical interactions between proteins by duplication, and review other mechanisms. We dissect the dataset of protein complexes of known three-dimensional structure, and show that roughly 90% of these complexes contain contacts between identical proteins within the same complex. Proteins that are shared across different complexes occur frequently, and they tend to be essential genes more often than members of a single protein complex. We also provide a perspective on the evolutionary mechanisms driving the growth of other modular cellular networks such as transcriptional regulatory and metabolic networks.
Figures



References
-
- Aharoni A, Gaidukov L, Khersonsky O, Mc Q.G.S, Roodveldt C, Tawfik D.S. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 2005;37:73–76. - PubMed
-
- Amoutzias G.D, Robertson D.L, Oliver S.G, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–279. 10.1038/sj.embor.7400096 - DOI - PMC - PubMed
-
- Amoutzias G.D, Weiner J, Bornberg-Bauer E. Phylogenetic profiling of protein interaction networks in eukaryotic transcription factors reveals focal proteins being ancestral to hubs. Gene. 2005;347:247–253. 10.1016/j.gene.2004.12.031 - DOI - PubMed
-
- Andreeva A, Howorth D, Brenner S.E, Hubbard T.J, Chothia C, Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. 10.1093/nar/gkh039 - DOI - PMC - PubMed
-
- Babu M.M, Luscombe N.M, Aravind L, Gerstein M, Teichmann S.A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. 10.1016/j.sbi.2004.05.004 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

