Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p
- PMID: 16524911
- PMCID: PMC1398067
- DOI: 10.1128/EC.5.3.568-578.2006
Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p
Abstract
Cox11p is an integral protein of the inner mitochondrial membrane that is essential for cytochrome c oxidase assembly. The bulk of the protein is located in the intermembrane space and displays high levels of evolutionary conservation. We have analyzed a collection of site-directed and random cox11 mutants in an effort to further define essential portions of the molecule. Of the alleles studied, more than half had no apparent effect on Cox11p function. Among the respiration deficiency-encoding alleles, we identified three distinct phenotypes, which included a set of mutants with a misassembled or partially assembled cytochrome oxidase, as indicated by a blue-shifted cytochrome aa(3) peak. In addition to the shifted spectral signal, these mutants also display a specific reduction in the levels of subunit 1 (Cox1p). Two of these mutations are likely to occlude a surface pocket behind the copper-binding domain in Cox11p, based on analogy with the Sinorhizobium meliloti Cox11 solution structure, thereby suggesting that this pocket is crucial for Cox11p function. Sequential deletions of the matrix portion of Cox11p suggest that this domain is not functional beyond the residues involved in mitochondrial targeting and membrane insertion. In addition, our studies indicate that Deltacox11, like Deltasco1, displays a specific hypersensitivity to hydrogen peroxide. Our studies provide the first evidence at the level of the cytochrome oxidase holoenzyme that Cox1p is the in vivo target for Cox11p and suggest that Cox11p may also have a role in the response to hydrogen peroxide exposure.
Figures

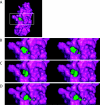
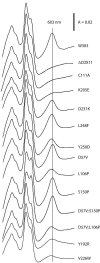


References
-
- Banci, L., I. Bertini, F. Cantini, S. Ciofi-Baffoni, L. Gonnelli, and S. Mangani. 2004. Solution structure of Cox11, a novel type of β-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J. Biol. Chem. 279:34833-34839. - PubMed
-
- Brazzolotto, X., J. Gaillard, K. Pantopoulos, M. W. Hentze, and J. M. Moulis. 1999. Human cytoplasmic aconitase (iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. J. Biol. Chem. 274:21625-21630. - PubMed
-
- Carr, H. S., G. N. George, and D. R. Winge. 2002. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 277:31237-31242. - PubMed
-
- Carr, H. S., A. B. Maxfield, Y.-C. Horng, and D. R. Winge. 2005. Functional analysis of the domains in Cox11. J. Biol. Chem. 280:22664-22669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

