Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa
- PMID: 16524913
- PMCID: PMC1398062
- DOI: 10.1128/EC.5.3.587-600.2006
Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa
Abstract
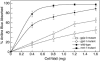
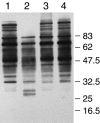
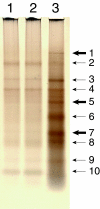
Using mutational and proteomic approaches, we have demonstrated the importance of the glycosylphosphatidylinositol (GPI) anchor pathway for cell wall synthesis and integrity and for the overall morphology of the filamentous fungus Neurospora crassa. Mutants affected in the gpig-1, gpip-1, gpip-2, gpip-3, and gpit-1 genes, which encode components of the N. crassa GPI anchor biosynthetic pathway, have been characterized. GPI anchor mutants exhibit colonial morphologies, significantly reduced rates of growth, altered hyphal growth patterns, considerable cellular lysis, and an abnormal "cell-within-a-cell" phenotype. The mutants are deficient in the production of GPI-anchored proteins, verifying the requirement of each altered gene for the process of GPI-anchoring. The mutant cell walls are abnormally weak, contain reduced amounts of protein, and have an altered carbohydrate composition. The mutant cell walls lack a number of GPI-anchored proteins, putatively involved in cell wall biogenesis and remodeling. From these studies, we conclude that the GPI anchor pathway is critical for proper cell wall structure and function in N. crassa.
Figures




References
-
- Alloush, H. M., J. L. Lopez-Ribot, B. J. Masten, and W. L. Chaffin. 1997. 3-Phosphoglycerate kinase: a glycolytic enzyme present in the cell wall of Candida albicans. Microbiology 143:321-330. - PubMed
-
- Angiolella, L., M. Facchin, A. Stringaro, B. Maras, N. Simonetti, and A. Cassone. 1996. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173:684-690. - PubMed
-
- Benachour, A., G. Sipos, I. Flury, F. Reggiori, E. Canivenc-Gansel, C. Vionnet, A. Conzelmann, and M. Benghezal. 1999. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 274:15251-15261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

