The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase
- PMID: 16524922
- PMCID: PMC1393255
- DOI: 10.1128/MMBR.70.1.177-191.2006
The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase
Abstract
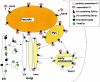
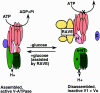
All eukaryotic cells contain multiple acidic organelles, and V-ATPases are central players in organelle acidification. Not only is the structure of V-ATPases highly conserved among eukaryotes, but there are also many regulatory mechanisms that are similar between fungi and higher eukaryotes. These mechanisms allow cells both to regulate the pHs of different compartments and to respond to changing extracellular conditions. The Saccharomyces cerevisiae V-ATPase has emerged as an important model for V-ATPase structure and function in all eukaryotic cells. This review discusses current knowledge of the structure, function, and regulation of the V-ATPase in S. cerevisiae and also examines the relationship between biosynthesis and transport of V-ATPase and compartment-specific regulation of acidification.
Figures

References
-
- Abazeed, M. E., J. M. Blanchette, and R. S. Fuller. 2004. Cell-free transport from the TGN to late endosome requires factors involved in formation and consumption of clathrin-coated vesicles. J. Biol. Chem. 280:4442-4450. - PubMed
-
- Arai, H., S. Pink, and M. Forgac. 1989. Interaction of anions and ATP with the coated vesicle proton pump. Biochemistry 28:3075-3082. - PubMed
-
- Arata, Y., J. D. Baleja, and M. Forgac. 2002. Localization of subunits D, E, and G in the yeast V-ATPase complex using cysteine-mediated cross-linking to subunit B. Biochemistry 41:11301-11307. - PubMed
-
- Bauerle, C., M. N. Ho, M. A. Lindorfer, and T. H. Stevens. 1993. The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H+-ATPase membrane sector. J. Biol. Chem. 268:12749-12757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

