Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria
- PMID: 16524923
- PMCID: PMC1393253
- DOI: 10.1128/MMBR.70.1.192-221.2006
Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria
Abstract
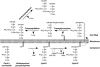
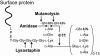
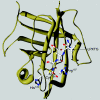
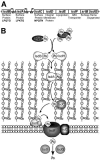
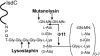
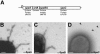
The cell wall envelopes of gram-positive bacteria represent a surface organelle that not only functions as a cytoskeletal element but also promotes interactions between bacteria and their environment. Cell wall peptidoglycan is covalently and noncovalently decorated with teichoic acids, polysaccharides, and proteins. The sum of these molecular decorations provides bacterial envelopes with species- and strain-specific properties that are ultimately responsible for bacterial virulence, interactions with host immune systems, and the development of disease symptoms or successful outcomes of infections. Surface proteins typically carry two topogenic sequences, i.e., N-terminal signal peptides and C-terminal sorting signals. Sortases catalyze a transpeptidation reaction by first cleaving a surface protein substrate at the cell wall sorting signal. The resulting acyl enzyme intermediates between sortases and their substrates are then resolved by the nucleophilic attack of amino groups, typically provided by the cell wall cross bridges of peptidoglycan precursors. The surface protein linked to peptidoglycan is then incorporated into the envelope and displayed on the microbial surface. This review focuses on the mechanisms of surface protein anchoring to the cell wall envelope by sortases and the role that these enzymes play in bacterial physiology and pathogenesis.
Figures

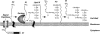
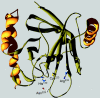
References
-
- Akabas, M. H., and A. Karlin. 1995. Identification of acetylcholine receptor channel-lining residues in the M1 segment of the alpha-subunit. Biochemistry 34:12496-12500. - PubMed
-
- Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190:727-738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

