Glucose signaling in Saccharomyces cerevisiae
- PMID: 16524925
- PMCID: PMC1393250
- DOI: 10.1128/MMBR.70.1.253-282.2006
Glucose signaling in Saccharomyces cerevisiae
Abstract
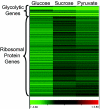
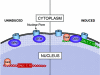
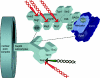
Eukaryotic cells possess an exquisitely interwoven and fine-tuned series of signal transduction mechanisms with which to sense and respond to the ubiquitous fermentable carbon source glucose. The budding yeast Saccharomyces cerevisiae has proven to be a fertile model system with which to identify glucose signaling factors, determine the relevant functional and physical interrelationships, and characterize the corresponding metabolic, transcriptomic, and proteomic readouts. The early events in glucose signaling appear to require both extracellular sensing by transmembrane proteins and intracellular sensing by G proteins. Intermediate steps involve cAMP-dependent stimulation of protein kinase A (PKA) as well as one or more redundant PKA-independent pathways. The final steps are mediated by a relatively small collection of transcriptional regulators that collaborate closely to maximize the cellular rates of energy generation and growth. Understanding the nuclear events in this process may necessitate the further elaboration of a new model for eukaryotic gene regulation, called "reverse recruitment." An essential feature of this idea is that fine-structure mapping of nuclear architecture will be required to understand the reception of regulatory signals that emanate from the plasma membrane and cytoplasm. Completion of this task should result in a much improved understanding of eukaryotic growth, differentiation, and carcinogenesis.
Figures


Similar articles
-
Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae.Sensors (Basel). 2010;10(6):6195-240. doi: 10.3390/s100606195. Epub 2010 Jun 21. Sensors (Basel). 2010. PMID: 22219709 Free PMC article. Review.
-
A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose.Mol Microbiol. 1999 Jun;32(5):1002-12. doi: 10.1046/j.1365-2958.1999.01413.x. Mol Microbiol. 1999. PMID: 10361302
-
Glucose- and nitrogen sensing and regulatory mechanisms in Saccharomyces cerevisiae.FEMS Yeast Res. 2014 Aug;14(5):683-96. doi: 10.1111/1567-1364.12157. Epub 2014 May 8. FEMS Yeast Res. 2014. PMID: 24738657 Review.
-
Glucose repression in Saccharomyces cerevisiae.FEMS Yeast Res. 2015 Sep;15(6):fov068. doi: 10.1093/femsyr/fov068. Epub 2015 Jul 22. FEMS Yeast Res. 2015. PMID: 26205245 Free PMC article. Review.
-
The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae.Genetics. 2005 Jul;170(3):1009-21. doi: 10.1534/genetics.105.043109. Epub 2005 May 6. Genetics. 2005. PMID: 15879503 Free PMC article.
Cited by
-
The Involvement of Mig1 from Xanthophyllomyces dendrorhous in Catabolic Repression: An Active Mechanism Contributing to the Regulation of Carotenoid Production.PLoS One. 2016 Sep 13;11(9):e0162838. doi: 10.1371/journal.pone.0162838. eCollection 2016. PLoS One. 2016. PMID: 27622474 Free PMC article.
-
Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae.PLoS Genet. 2016 Oct 14;12(10):e1006372. doi: 10.1371/journal.pgen.1006372. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27741250 Free PMC article.
-
Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae.Eukaryot Cell. 2012 Feb;11(2):119-28. doi: 10.1128/EC.05061-11. Epub 2011 Dec 2. Eukaryot Cell. 2012. PMID: 22140226 Free PMC article.
-
The glucose sensor-like protein Hxs1 is a high-affinity glucose transporter and required for virulence in Cryptococcus neoformans.PLoS One. 2013 May 14;8(5):e64239. doi: 10.1371/journal.pone.0064239. Print 2013. PLoS One. 2013. PMID: 23691177 Free PMC article.
-
A negative feedback loop at the nuclear periphery regulates GAL gene expression.Mol Biol Cell. 2012 Apr;23(7):1367-75. doi: 10.1091/mbc.E11-06-0547. Epub 2012 Feb 9. Mol Biol Cell. 2012. PMID: 22323286 Free PMC article.
References
-
- Ahn, J. H., S. H. Park, and H. S. Kang. 1995. Inactivation of the UAS1 of STA1 by glucose and STA10 and identification of two loci, SNS1 and MSS1, involved in STA10-dependent repression in Saccharomyces cerevisiae. Mol. Gen. Genet. 246:529-537. - PubMed
-
- Ahuatzi, D., P. Herrero, T. de la Cera, and F. Moreno. 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279:14440-14446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

