Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY
- PMID: 16525509
- PMCID: PMC1422171
- DOI: 10.1038/sj.emboj.7601028
Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY
Erratum in
- EMBO J. 2006 May 3;25(9):2038
Abstract
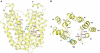
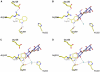
Cation-coupled active transport is an essential cellular process found ubiquitously in all living organisms. Here, we present two novel ligand-free X-ray structures of the lactose permease (LacY) of Escherichia coli determined at acidic and neutral pH, and propose a model for the mechanism of coupling between lactose and H+ translocation. No sugar-binding site is observed in the absence of ligand, and deprotonation of the key residue Glu269 is associated with ligand binding. Thus, substrate induces formation of the sugar-binding site, as well as the initial step in H+ transduction.
Figures
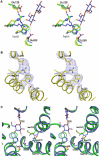


References
-
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301: 610–615 - PubMed
-
- Brünger A, Adams P, Clore G, DeLano W, Gros P, Grosse-Kunstleve R, Jiang J, Kuszewski J, Nilges M, Pannu N, Read R, Rice L, Simonson T, Warren G (1998) Crystallography & NMR Systems: a new software suite for macromolecular structure determination. Act Crystallogr D54: 905–921 - PubMed
-
- Frillingos S, Sahin-Toth M, Wu J, Kaback HR (1998) Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J 12: 1281–1299 - PubMed
-
- Guan L, Hu Y, Kaback HR (2003) Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry 42: 1377–1382 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

