Sp1/3 and NF-1 mediate basal transcription of the human P2X1 gene in megakaryoblastic MEG-01 cells
- PMID: 16529657
- PMCID: PMC1464135
- DOI: 10.1186/1471-2199-7-10
Sp1/3 and NF-1 mediate basal transcription of the human P2X1 gene in megakaryoblastic MEG-01 cells
Abstract
Background: P2X1 receptors play an important role in platelet function as they can induce shape change, granule centralization and are also involved in thrombus formation. As platelets have no nuclei, the level of P2X1 expression depends on transcriptional regulation in megakaryocytes, the platelet precursor cell. Since nothing is known about the molecular mechanisms regulating megakaryocytic P2X1 expression, this study aimed to identify and functionally characterize the P2X1 core promoter utilized in the human megakaryoblastic cell line MEG-01.
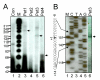
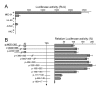
Results: In order to identify cis-acting elements involved in the transcriptional regulation of P2X1 expression, the ability of 4.7 kb P2X1 upstream sequence to drive luciferase reporter gene expression was tested. Low promoter activity was detected in proliferating MEG-01 cells. This activity increased 20-fold after phorbol-12-myristate-13-acetate (PMA) induced differentiation. A transcription start site was detected 365 bp upstream of the start codon by primer extension. Deletion analysis of reporter constructs indicated a core promoter located within the region -68 to +149 bp that contained two Sp1 sites (named Sp1a and Sp1b) and an NF-1 site. Individual mutations of Sp1b or NF-1 binding sites severely reduced promoter activity whereas triple mutation of Sp1a, Sp1b and NF-1 sites completely abolished promoter activity in both untreated and PMA treated cells. Sp1/3 and NF-1 proteins were shown to bind their respective sites by EMSA and interaction of Sp1/3, NF-1 and TFIIB with the endogenous P2X1 core promoter in MEG-01 cells was demonstrated by chromatin immunoprecipitation. Alignment of P2X1 genes from human, chimp, rat, mouse and dog revealed consensus Sp1a, Sp1b and NF-1 binding sites in equivalent positions thereby demonstrating evolutionary conservation of these functionally important sites.
Conclusion: This study has identified and characterized the P2X1 promoter utilized in MEG-01 cells and shown that binding of Sp1/3 and NF-1 to elements in the direct vicinity of the transcription start site is essential for basal transcription. Targeting the function of these transcription factors in megakaryocytes may therefore provide a basis for the future therapeutic manipulation of platelet P2X1 function.
Figures





References
-
- North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

