Highly specific gene silencing by artificial microRNAs in Arabidopsis
- PMID: 16531494
- PMCID: PMC1456875
- DOI: 10.1105/tpc.105.039834
Highly specific gene silencing by artificial microRNAs in Arabidopsis
Abstract
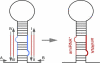
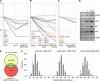
Plant microRNAs (miRNAs) affect only a small number of targets with high sequence complementarity, while animal miRNAs usually have hundreds of targets with limited complementarity. We used artificial miRNAs (amiRNAs) to determine whether the narrow action spectrum of natural plant miRNAs reflects only intrinsic properties of the plant miRNA machinery or whether it is also due to past selection against natural miRNAs with broader specificity. amiRNAs were designed to target individual genes or groups of endogenous genes. Like natural miRNAs, they had varying numbers of target mismatches. Previously determined parameters of target selection for natural miRNAs could accurately predict direct targets of amiRNAs. The specificity of amiRNAs, as deduced from genome-wide expression profiling, was as high as that of natural plant miRNAs, supporting the notion that extensive base pairing with targets is required for plant miRNA function. amiRNAs make an effective tool for specific gene silencing in plants, especially when several related, but not identical, target genes need to be downregulated. We demonstrate that amiRNAs are also active when expressed under tissue-specific or inducible promoters, with limited nonautonomous effects. The design principles for amiRNAs have been generalized and integrated into a Web-based tool (http://wmd.weigelworld.org).
Figures



References
-
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., and Pasquinelli, A.E. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122 553–563. - PubMed
-
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

