Spread of an inactive form of caspase-12 in humans is due to recent positive selection
- PMID: 16532395
- PMCID: PMC1424700
- DOI: 10.1086/503116
Spread of an inactive form of caspase-12 in humans is due to recent positive selection
Abstract
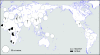
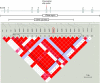
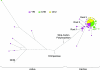
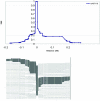
The human caspase-12 gene is polymorphic for the presence or absence of a stop codon, which results in the occurrence of both active (ancestral) and inactive (derived) forms of the gene in the population. It has been shown elsewhere that carriers of the inactive gene are more resistant to severe sepsis. We have now investigated whether the inactive form has spread because of neutral drift or positive selection. We determined its distribution in a worldwide sample of 52 populations and resequenced the gene in 77 individuals from the HapMap Yoruba, Han Chinese, and European populations. There is strong evidence of positive selection from low diversity, skewed allele-frequency spectra, and the predominance of a single haplotype. We suggest that the inactive form of the gene arose in Africa approximately 100-500 thousand years ago (KYA) and was initially neutral or almost neutral but that positive selection beginning approximately 60-100 KYA drove it to near fixation. We further propose that its selective advantage was sepsis resistance in populations that experienced more infectious diseases as population sizes and densities increased.
Figures

References
Web Resources
-
- DnaSP, http://www.ub.es/dnasp/
-
- Fluxus, http://www.fluxus-technology.com/ (for Network4.1.0.9)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the CASP12 genomic DNA sequence [accession number NC_000011] and the chimpanzee CASP12 sequence [accession number NW_113990])
References
-
- Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

