Recently mobilized transposons in the human and chimpanzee genomes
- PMID: 16532396
- PMCID: PMC1424692
- DOI: 10.1086/501028
Recently mobilized transposons in the human and chimpanzee genomes
Abstract
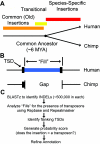
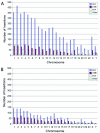
Transposable genetic elements are abundant in the genomes of most organisms, including humans. These endogenous mutagens can alter genes, promote genomic rearrangements, and may help to drive the speciation of organisms. In this study, we identified almost 11,000 transposon copies that are differentially present in the human and chimpanzee genomes. Most of these transposon copies were mobilized after the existence of a common ancestor of humans and chimpanzees, approximately 6 million years ago. Alu, L1, and SVA insertions accounted for >95% of the insertions in both species. Our data indicate that humans have supported higher levels of transposition than have chimpanzees during the past several million years and have amplified different transposon subfamilies. In both species, approximately 34% of the insertions were located within known genes. These insertions represent a form of species-specific genetic variation that may have contributed to the differential evolution of humans and chimpanzees. In addition to providing an initial overview of recently mobilized elements, our collections will be useful for assessing the impact of these insertions on their hosts and for studying the transposition mechanisms of these elements.
Figures

References
-
- Boissinot S, Chevret P, Furano A (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 - PubMed
-
- Boissinot S, Entezam A, Furano AV (2001) Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 18:926–935 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

