Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus
- PMID: 16533886
- PMCID: PMC2118231
- DOI: 10.1084/jem.20052116
Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus
Abstract
The predictability of virus-host interactions and disease progression in rapidly evolving human viral infections has been difficult to assess because of host and genetic viral diversity. Here we examined adaptive HIV-specific cellular and humoral immune responses and viral evolution in adult monozygotic twins simultaneously infected with the same virus. CD4 T cell counts and viral loads followed similar trajectories over three years of follow up. The initial CD8 T cell response targeted 17 epitopes, 15 of which were identical in each twin, including two immunodominant responses. By 36 months after infection, 14 of 15 initial responses were still detectable in both, whereas all new responses were subdominant and remained so. Of four responses that declined in both twins, three demonstrated mutations at the same residue. In addition, the evolving antibody responses cross-neutralized the other twin's virus, with similar changes in the pattern of evolution in the envelope gene. These results reveal considerable concordance of adaptive cellular and humoral immune responses and HIV evolution in the same genetic environment, suggesting constraints on mutational pathways to HIV immune escape.
Figures

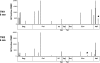
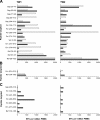





Comment in
-
Immune responses and HIV: a little order from the chaos.J Exp Med. 2006 Mar 20;203(3):501-3. doi: 10.1084/jem.20060216. Epub 2006 Mar 13. J Exp Med. 2006. PMID: 16533889 Free PMC article.
References
-
- Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B.H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science. 296:2354–2360. - PubMed
-
- Preston, B.D., B.J. Poiesz, and L.A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science. 242:1168–1171. - PubMed
-
- Wei, X., S.K. Ghosh, M.E. Taylor, V.A. Johnson, E.A. Emini, P. Deutsch, J.D. Lifson, S. Bonhoeffer, M.A. Nowak, B.H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 373:117–122. - PubMed
-
- Ho, D.D., A.U. Neumann, A.S. Perelson, W. Chen, J.M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 373:123–126. - PubMed
-
- Coffin, J.M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 267:483–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

