Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression
- PMID: 16533945
- PMCID: PMC2063729
- DOI: 10.1083/jcb.200506119
Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression
Abstract
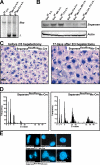
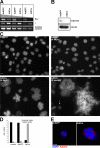
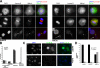
Separase is a protease whose liberation from its inhibitory chaperone Securin triggers sister chromatid disjunction at anaphase onset in yeast by cleaving cohesin's kleisin subunit. We have created conditional knockout alleles of the mouse Separase and Securin genes. Deletion of both copies of Separase but not Securin causes embryonic lethality. Loss of Securin reduces Separase activity because deletion of just one copy of the Separase gene is lethal to embryos lacking Securin. In embryonic fibroblasts, Separase depletion blocks sister chromatid separation but does not prevent other aspects of mitosis, cytokinesis, or chromosome replication. Thus, fibroblasts lacking Separase become highly polyploid. Hepatocytes stimulated to proliferate in vivo by hepatectomy also become unusually large and polyploid in the absence of Separase but are able to regenerate functional livers. Separase depletion in bone marrow causes aplasia and the presumed death of hematopoietic cells other than erythrocytes. Destruction of sister chromatid cohesion by Separase may be a universal feature of mitosis in eukaryotic cells.
Figures





References
-
- Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 93:1067–1076. - PubMed
-
- Cohen-Fix, O., J.-M. Peters, M.W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081–3093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

