The molecular elasticity of the insect flight muscle proteins projectin and kettin
- PMID: 16537423
- PMCID: PMC1450192
- DOI: 10.1073/pnas.0509016103
The molecular elasticity of the insect flight muscle proteins projectin and kettin
Abstract
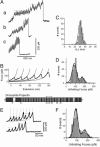
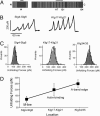
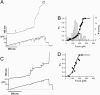
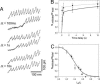
Projectin and kettin are titin-like proteins mainly responsible for the high passive stiffness of insect indirect flight muscles, which is needed to generate oscillatory work during flight. Here we report the mechanical properties of kettin and projectin by single-molecule force spectroscopy. Force-extension and force-clamp curves obtained from Lethocerus projectin and Drosophila recombinant projectin or kettin fragments revealed that fibronectin type III domains in projectin are mechanically weaker (unfolding force, F(u) approximately 50-150 pN) than Ig-domains (F(u) approximately 150-250 pN). Among Ig domains in Sls/kettin, the domains near the N terminus are less stable than those near the C terminus. Projectin domains refolded very fast [85% at 15 s(-1) (25 degrees C)] and even under high forces (15-30 pN). Temperature affected the unfolding forces with a Q(10) of 1.3, whereas the refolding speed had a Q(10) of 2-3, probably reflecting the cooperative nature of the folding mechanism. High bending rigidities of projectin and kettin indicated that straightening the proteins requires low forces. Our results suggest that titin-like proteins in indirect flight muscles could function according to a folding-based-spring mechanism.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Josephson R. K., Malamud J. G., Stokes D. R. J. Exp. Biol. 2000;203:2713–2722. - PubMed
-
- Pringle J. W. Proc. R. Soc. London Ser. B; 1978. pp. 107–130. - PubMed
-
- Bullard B., Leake M. C., Leonard K. In: Nature's Versatile Engine: Insect Flight Muscle Inside and Out. Vigoreaux J., editor. Georgetown, TX: Landes Bioscience; 2004. pp. 177–186.
-
- Bullard B., Leonard K. Adv. Biophys. 1996;33:211–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

