Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray
- PMID: 16537477
- PMCID: PMC1449637
- DOI: 10.1073/pnas.0510921103
Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray
Abstract
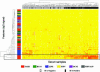
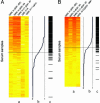
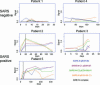
To monitor severe acute respiratory syndrome (SARS) infection, a coronavirus protein microarray that harbors proteins from SARS coronavirus (SARS-CoV) and five additional coronaviruses was constructed. These microarrays were used to screen approximately 400 Canadian sera from the SARS outbreak, including samples from confirmed SARS-CoV cases, respiratory illness patients, and healthcare professionals. A computer algorithm that uses multiple classifiers to predict samples from SARS patients was developed and used to predict 206 sera from Chinese fever patients. The test assigned patients into two distinct groups: those with antibodies to SARS-CoV and those without. The microarray also identified patients with sera reactive against other coronavirus proteins. Our results correlated well with an indirect immunofluorescence test and demonstrated that viral infection can be monitored for many months after infection. We show that protein microarrays can serve as a rapid, sensitive, and simple tool for large-scale identification of viral-specific antibodies in sera.
Conflict of interest statement
Conflict of interest statement: M.S. consults for Invitrogen.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

