Rapid, sequence-specific detection of unpurified PCR amplicons via a reusable, electrochemical sensor
- PMID: 16537478
- PMCID: PMC1449638
- DOI: 10.1073/pnas.0511325103
Rapid, sequence-specific detection of unpurified PCR amplicons via a reusable, electrochemical sensor
Abstract
We report an electrochemical method for the sequence-specific detection of unpurified amplification products of the gyrB gene of Salmonella typhimurium. Using an asymmetric PCR and the electrochemical E-DNA detection scheme, single-stranded amplicons were produced from as few as 90 gene copies and, without subsequent purification, rapidly identified. The detection is specific; the sensor does not respond when challenged with control oligonucleotides based on the gyrB genes of either Escherichia coli or various Shigella species. In contrast to existing sequence-specific optical- and capillary electrophoresis-based detection methods, the E-DNA sensor is fully electronic and requires neither cumbersome, expensive optics nor high voltage power supplies. Given these advantages, E-DNA sensors appear well suited for implementation in portable PCR microdevices directed at, for example, the rapid detection of pathogens.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


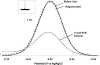
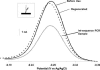

Similar articles
-
[Detection of the determinants of tetracycline A, B and C resistance in Shigella and Salmonella using DNA probes].Antibiot Khimioter. 1991 Jul;36(7):17-9. Antibiot Khimioter. 1991. PMID: 1953181 Russian.
-
Molecular characterization of intact, but cryptic, flagellin genes in the genus Shigella.Mol Microbiol. 1994 Apr;12(2):277-85. doi: 10.1111/j.1365-2958.1994.tb01016.x. Mol Microbiol. 1994. PMID: 8057852
-
The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island.Mol Microbiol. 1999 Jul;33(1):63-73. doi: 10.1046/j.1365-2958.1999.01448.x. Mol Microbiol. 1999. PMID: 10411724
-
Electrochemical genosensor based on peptide nucleic acid-mediated PCR and asymmetric PCR techniques: Electrostatic interactions with a metal cation.Anal Chem. 2006 Apr 1;78(7):2182-9. doi: 10.1021/ac051526a. Anal Chem. 2006. PMID: 16579596
-
Genetic determinants of virulence in Shigella and dysenteric strains of Escherichia coli: their involvement in the pathogenesis of dysentery.Curr Top Microbiol Immunol. 1985;118:71-95. doi: 10.1007/978-3-642-70586-1_5. Curr Top Microbiol Immunol. 1985. PMID: 2414072 Review. No abstract available.
Cited by
-
Detection of single-nucleotide polymorphism on uidA gene of Escherichia coli by a multiplexed electrochemical DNA biosensor with oligonucleotide-incorporated nonfouling surface.Sensors (Basel). 2011;11(8):8018-27. doi: 10.3390/s110808018. Epub 2011 Aug 15. Sensors (Basel). 2011. PMID: 22164059 Free PMC article.
-
Monitoring cooperative binding using electrochemical DNA-based sensors.Langmuir. 2015 Jan 20;31(2):868-75. doi: 10.1021/la504083c. Epub 2015 Jan 6. Langmuir. 2015. PMID: 25517392 Free PMC article.
-
Strategies for the preparation of non-amplified and amplified genomic dengue gene samples for electrochemical DNA biosensing applications.RSC Adv. 2021 Dec 20;12(1):1-10. doi: 10.1039/d1ra06753b. eCollection 2021 Dec 20. RSC Adv. 2021. PMID: 35424522 Free PMC article.
-
Electrochemical genosensor for specific detection of the food-borne pathogen, Vibrio cholerae.World J Microbiol Biotechnol. 2012 Apr;28(4):1699-706. doi: 10.1007/s11274-011-0978-x. Epub 2011 Dec 18. World J Microbiol Biotechnol. 2012. PMID: 22805952
-
Design, synthesis, and characterization of nucleic-acid-functionalized gold surfaces for biomarker detection.Langmuir. 2012 Jan 17;28(2):1068-82. doi: 10.1021/la2028862. Epub 2011 Sep 28. Langmuir. 2012. PMID: 21905721 Free PMC article.
References
-
- Tappero J. W., Schuchat A., Deaver K. A., Mascola L., Wenger J. D. J. Am. Med. Soc. 1995;273:1118–1122. - PubMed
-
- Bopp C. A., Brenner F. W., Fields P. I., Wells J. G., Strockbine N. A. In: Manual of Clinical Microbiology. Murray P. R., Braron E. J., Jorgensen J. H., Pfaller M. A., Yolken R. H., editors. Washington, DC: Am. Soc. Microbiol.; 2003. pp. 654–671.
-
- Boyce T. G., Swerdlow D. L., Griffin P. M. N. Engl. J. Med. 1995;333:364–368. - PubMed
-
- United States Department of Agriculture Animal and Plant Health Inspection Services . The national poultry improvement plan. Subasesart B. Bacteriological examination procedure 147–11. Laboratory procedure recommended for the bacteriological examination of Salmonella. Washington, DC: U.S. Depart. Agric.; 1996. pp. 14–19.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

