Structural basis of hepatocyte growth factor/scatter factor and MET signalling
- PMID: 16537482
- PMCID: PMC1449643
- DOI: 10.1073/pnas.0509040103
Structural basis of hepatocyte growth factor/scatter factor and MET signalling
Abstract
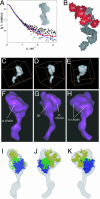
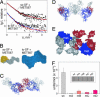
The polypeptide growth factor, hepatocyte growth factor/scatter factor (HGF/SF), shares the multidomain structure and proteolytic mechanism of activation of plasminogen and other complex serine proteinases. HGF/SF, however, has no enzymatic activity. Instead, it controls the growth, morphogenesis, or migration of epithelial, endothelial, and muscle progenitor cells through the receptor tyrosine kinase MET. Using small-angle x-ray scattering and cryo-electron microscopy, we show that conversion of pro(single-chain)HGF/SF into the active two-chain form is associated with a major structural transition from a compact, closed conformation to an elongated, open one. We also report the structure of a complex between two-chain HGF/SF and the MET ectodomain (MET928) with 1:1 stoichiometry in which the N-terminal and first kringle domain of HGF/SF contact the face of the seven-blade beta-propeller domain of MET harboring the loops connecting the beta-strands b-c and d-a, whereas the C-terminal serine proteinase homology domain binds the opposite "b" face. Finally, we describe a complex with 2:2 stoichiometry between two-chain HGF/SF and a truncated form of the MET ectodomain (MET567), which is assembled around the dimerization interface seen in the crystal structure of the NK1 fragment of HGF/SF and displays the features of a functional, signaling unit. The study shows how the proteolytic mechanism of activation of the complex proteinases has been adapted to cell signaling in vertebrate organisms, offers a description of monomeric and dimeric ligand-receptor complexes, and provides a foundation to the structural basis of HGF/SF-MET signaling.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Nakamura T., Nishizawa T., Hagiya M., Seki T., Shimonishi M., Sugimura A., Tashiro K., Shimizu S. Nature. 1989;342:440–443. - PubMed
-
- Miyazawa K., Tsubouchi H., Naka D., Takahashi K., Okigaki M., Arakaki N., Nakayama H., Hirono S., Sakiyama O., Takahashi K., et al. Biochem. Biophys. Res. Commun. 1989;163:967–973. - PubMed
-
- Stoker M., Gherardi E., Perryman M., Gray J. Nature. 1987;327:239–242. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

