Mutagenesis analysis of the nsp4 main proteinase reveals determinants of arterivirus replicase polyprotein autoprocessing
- PMID: 16537610
- PMCID: PMC1440411
- DOI: 10.1128/JVI.80.7.3428-3437.2006
Mutagenesis analysis of the nsp4 main proteinase reveals determinants of arterivirus replicase polyprotein autoprocessing
Abstract
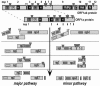
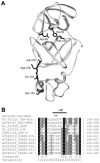
Nonstructural protein 4 (nsp4; 204 amino acids) is the chymotrypsin-like serine main proteinase of the arterivirus Equine arteritis virus (order Nidovirales), which controls the maturation of the replicase complex. nsp4 includes a unique C-terminal domain (CTD) connected to the catalytic two-beta-barrel structure by the poorly conserved residues 155 and 156. This dipeptide might be part of a hinge region (HR) that facilitates interdomain movements and thereby regulates (in time and space) autoprocessing of replicase polyproteins pp1a and pp1ab at eight sites that are conserved in arteriviruses. To test this hypothesis, we characterized nsp4 proteinase mutants carrying either point mutations in the putative HR domain or a large deletion in the CTD. When tested in a reverse genetics system, three groups of mutants were recognized (wild-type-like, debilitated, and dead), which was in line with the expected impact of mutations on HR flexibility. When tested in a transient expression system, the effects of the mutations on the production and turnover of replicase proteins varied widely. They were cleavage product specific and revealed a pronounced modulating effect of moieties derived from the nsp1-3 region of pp1a. Mutations that were lethal affected the efficiency of polyprotein autoprocessing most strongly. These mutants may be impaired in the accumulation of nsp5-7 and/or suffer from delayed or otherwise perturbed processing at the nsp5/6 and nsp6/7 junctions. On average, the production of nsp7-8 seems to be the most resistant to debilitating nsp4 mutations. Our results further prove that the CTD is essential for a vital nsp4 property other than catalysis.
Figures




Similar articles
-
Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7.J Gen Virol. 2006 Dec;87(Pt 12):3473-3482. doi: 10.1099/vir.0.82269-0. J Gen Virol. 2006. PMID: 17098961
-
Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease.J Virol. 1997 Dec;71(12):9313-22. doi: 10.1128/JVI.71.12.9313-9322.1997. J Virol. 1997. PMID: 9371590 Free PMC article.
-
The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases.J Biol Chem. 1996 Mar 1;271(9):4864-71. doi: 10.1074/jbc.271.9.4864. J Biol Chem. 1996. PMID: 8617757
-
Adaptive Mutations in Replicase Transmembrane Subunits Can Counteract Inhibition of Equine Arteritis Virus RNA Synthesis by Cyclophilin Inhibitors.J Virol. 2019 Aug 28;93(18):e00490-19. doi: 10.1128/JVI.00490-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31243130 Free PMC article.
-
The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins.Virus Res. 2010 Dec;154(1-2):61-76. doi: 10.1016/j.virusres.2010.07.030. Epub 2010 Aug 7. Virus Res. 2010. PMID: 20696193 Free PMC article. Review.
Cited by
-
Equine arteritis virus.Vet Microbiol. 2013 Nov 29;167(1-2):93-122. doi: 10.1016/j.vetmic.2013.06.015. Epub 2013 Jul 3. Vet Microbiol. 2013. PMID: 23891306 Free PMC article. Review.
-
Experiences with infectious cDNA clones of equine arteritis virus: lessons learned and insights gained.Virology. 2014 Aug;462-463:388-403. doi: 10.1016/j.virol.2014.04.029. Epub 2014 Jun 7. Virology. 2014. PMID: 24913633 Free PMC article. Review.
-
Aspartic acid at residue 185 modulates the capacity of HP-PRRSV nsp4 to antagonize IFN-I expression.Virology. 2020 Jul;546:79-87. doi: 10.1016/j.virol.2020.04.007. Epub 2020 Apr 21. Virology. 2020. PMID: 32452419 Free PMC article.
-
Structure and genetic analysis of the arterivirus nonstructural protein 7alpha.J Virol. 2011 Jul;85(14):7449-53. doi: 10.1128/JVI.00255-11. Epub 2011 May 11. J Virol. 2011. PMID: 21561912 Free PMC article.
-
Enhanced Porcine Reproductive and Respiratory Syndrome Virus Replication in Nsp4- or Nsp2-Overexpressed Marc-145 Cell Lines.Vet Sci. 2025 Jan 13;12(1):52. doi: 10.3390/vetsci12010052. Vet Sci. 2025. PMID: 39852927 Free PMC article.
References
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
-
- Balasuriya, U. B., J. F. Hedges, V. L. Smalley, A. Navarrette, W. H. McCollum, P. J. Timoney, E. J. Snijder, and N. J. MacLachlan. 2004. Genetic characterization of equine arteritis virus during persistent infection of stallions. J. Gen. Virol. 85:379-390. - PubMed
-
- Barrette-Ng, I. H., K. K. S. Ng, B. L. Mark, D. van Aken, M. M. Cherney, C. Garen, Y. Kolodenko, A. E. Gorbalenya, E. J. Snijder, and M. N. G. James. 2002. Structure of arterivirus nsp4—the smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 277:39960-39966. - PubMed
-
- Bryans, J. T., E. R. Doll, and R. E. Knappenberger. 1957. An outbreak of abortion caused by the equine arteritis virus. Cornell Vet. 47:69-75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

