Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen modulates K1 expression through its cis-acting elements within the terminal repeats
- PMID: 16537612
- PMCID: PMC1440413
- DOI: 10.1128/JVI.80.7.3445-3458.2006
Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen modulates K1 expression through its cis-acting elements within the terminal repeats
Abstract
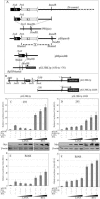
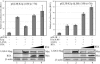
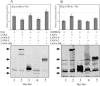
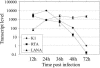
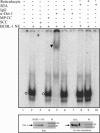
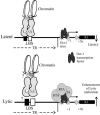
K1 is the first open reading frame encoded by Kaposi's sarcoma-associated herpesvirus (KSHV) and lies positionally to the immediate right of the terminal repeats. K1 is a transmembrane glycoprotein having a functional immunoreceptor tyrosine-based activation motif (ITAM) capable of activating B-cell receptor signaling. K1 is expressed mostly during the lytic cycle of the virus and its promoter lies within the terminal repeat which contains the binding sites for latency-associated nuclear antigen (LANA). The K1 promoter (K1p) having LANA binding sites assayed by reporter assay demonstrated that LANA is capable of down-regulating K1 promoter transcriptional activity. However, the KSHV replication transcription activator RTA up-regulates K1p transcriptional activity. The promoter deleted of LANA binding sites showed loss in LANA-mediated down-regulation but was unaffected for RTA-mediated up-regulation. Increasing amounts of RTA rescued LANA-mediated repression of K1p transcriptional activity in cotransfection experiments. Reporter assay data suggest that LANA binding to its cognate sequence is critical for LANA-mediated repression of K1p as a LANA construct lacking the DNA binding domain was unable to repress K1p transcription. Additionally, KSHV primary infection experiments suggest that K1 is expressed during early infection but is repressed on the establishment of latency and so follows an expression profile similar to that of RTA during infection. Analysis of the promoter sequence revealed the presence of Oct-1 transcription factor binding sites within the -116 to +76 region. Mutational analysis of the Oct-1 sites abolished RTA-mediated transcriptional activation, suggesting that RTA up-regulates K1p transcription through binding to this transcription factor.
Figures




Similar articles
-
Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency.J Virol. 2005 Jun;79(12):7453-65. doi: 10.1128/JVI.79.12.7453-7465.2005. J Virol. 2005. PMID: 15919901 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency.J Virol. 2004 Jun;78(12):6585-94. doi: 10.1128/JVI.78.12.6585-6594.2004. J Virol. 2004. PMID: 15163750 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway.J Virol. 2005 Mar;79(6):3468-78. doi: 10.1128/JVI.79.6.3468-3478.2005. J Virol. 2005. PMID: 15731241 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
Cited by
-
Murine gammaherpesvirus 68 LANA acts on terminal repeat DNA to mediate episome persistence.J Virol. 2012 Nov;86(21):11863-76. doi: 10.1128/JVI.01656-12. Epub 2012 Aug 22. J Virol. 2012. PMID: 22915819 Free PMC article.
-
Characterization of de novo lytic infection of dermal lymphatic microvascular endothelial cells by Kaposi's sarcoma-associated herpesvirus.Virology. 2019 Oct;536:27-31. doi: 10.1016/j.virol.2019.07.028. Epub 2019 Jul 31. Virology. 2019. PMID: 31394409 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA contributes to viral latent replication by activating phosphorylation of survivin.J Virol. 2014 Apr;88(8):4204-17. doi: 10.1128/JVI.03855-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478433 Free PMC article.
-
Single molecule analysis of replicated DNA reveals the usage of multiple KSHV genome regions for latent replication.PLoS Pathog. 2011 Nov;7(11):e1002365. doi: 10.1371/journal.ppat.1002365. Epub 2011 Nov 3. PLoS Pathog. 2011. PMID: 22072974 Free PMC article.
-
mRNA translation regulation by the Gly-Ala repeat of Epstein-Barr virus nuclear antigen 1.J Virol. 2009 Feb;83(3):1289-98. doi: 10.1128/JVI.01369-08. Epub 2008 Nov 19. J Virol. 2009. PMID: 19019958 Free PMC article.
References
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
-
- Boshoff, C., D. Whitby, T. Hatziioannou, C. Fisher, J. van der Walt, A. Hatzakis, R. Weiss, and T. Schulz. 1995. Kaposi's-sarcoma-associated herpesvirus in HIV-negative Kaposi's sarcoma. Lancet 345:1043-1044. - PubMed
-
- Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

