Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses
- PMID: 16537839
- PMCID: PMC1401513
- DOI: 10.1093/nar/gkl054
Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses
Abstract
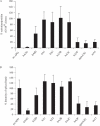
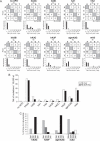
We demonstrated previously that the cytosine deaminase APOBEC3G inhibits retrotransposition of two active murine endogenous retroviruses, namely intracisternal A-particles (IAP) and MusD, in an ex vivo assay where retrotransposition was monitored by selection of neo-marked elements. Sequencing of the transposed copies further disclosed extensive editing, resulting in a high load of G-to-A mutations. Here, we asked whether this G-to-A editing was associated with an impact of APOBEC3G on viral cDNA yields. To this end, we used a specially designed quantitative PCR method to selectively measure the copy number of transposed retroelements, in the absence of G418 selection. We show that human APOBEC3G severely reduces the number of MusD and IAP transposed cDNA copies, with no effect on the level of the intermediate RNA transcripts. The magnitude of the decrease closely parallels that observed when transposed copies are assayed by selection of G418-resistant cells. Moreover, sequencing of transposed elements recovered by PCR without prior selection of the cells reveals high-level editing. Using this direct method with a series of cytosine deaminases, we further demonstrate a similar dual effect of African green monkey APOBE3G, human APOBEC3F and murine APOBEC3 on MusD retrotransposition, with a distinct extent and site specificity for each editing activity. Altogether the data demonstrate that cytosine deaminases have a protective effect against endogenous retroviruses both by reducing viral cDNA levels and by introducing mutations in the transposed copies, thus inactivating them for subsequent rounds of retrotransposition. This dual, two-step effect likely participates in the efficient defense of the cell genome against invading endogenous retroelements.
Figures


References
-
- Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H., Jr Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. - PubMed
-
- Boeke J.D., Stoye J.P. In: Retroviruses. Coffin J.M., Hughes S.H., Varmus H.E., editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. - PubMed
-
- Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Löwer R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. 1999;7:350–356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

