Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex
- PMID: 16537910
- PMCID: PMC1430322
- DOI: 10.1128/MCB.26.7.2661-2674.2006
Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex
Abstract
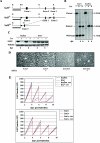
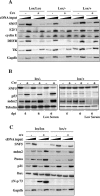
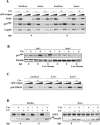
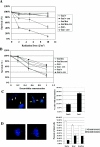
The gene encoding the SNF5/Ini1 core subunit of the SWI/SNF chromatin remodeling complex is a tumor suppressor in humans and mice, with an essential role in early embryonic development. To investigate further the function of this gene, we have generated a Cre/lox-conditional mouse line. We demonstrate that Snf5 deletion in primary fibroblasts impairs cell proliferation and survival without the expected derepression of most retinoblastoma protein-controlled, E2F-responsive genes. Furthermore, Snf5-deficient cells are hypersensitive to genotoxic stress, display increased aberrant mitotic features, and accumulate phosphorylated p53, leading to elevated expression of a specific subset of p53 target genes, suggesting a role for Snf5 in the DNA damage response. p53 inactivation does not rescue the proliferation defect caused by Snf5 deficiency but reduces apoptosis and strongly accelerates tumor formation in Snf5-heterozygous mice.
Figures


References
-
- Ameyar-Zazoua, M., M. B. Wisniewska, L. Bakiri, E. F. Wagner, M. Yaniv, and J. B. Weitzman. 2005. AP-1 dimers regulate transcription of the p14/p19ARF tumor suppressor gene. Oncogene 24:2298-2306. - PubMed
-
- Andreassen, P. R., F. B. Lacroix, O. D. Lohez, and R. L. Margolis. 2001. Neither p21WAF1 nor 14-3-3sigma prevents G2 progression to mitotic catastrophe in human colon carcinoma cells after DNA damage, but p21WAF1 induces stable G1 arrest in resulting tetraploid cells. Cancer Res. 61:7660-7668. - PubMed
-
- Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. - PubMed
-
- Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
