Enantiomer discrimination illustrated by the high resolution crystal structures of type 4 phosphodiesterase
- PMID: 16539372
- PMCID: PMC2527038
- DOI: 10.1021/jm051273d
Enantiomer discrimination illustrated by the high resolution crystal structures of type 4 phosphodiesterase
Abstract
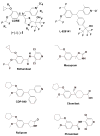
Type 4 phosphodiesterase (PDE4) inhibitors are emerging as new treatments for a number of disorders including asthma and chronic obstructive pulmonary disease. Here we report the biochemical characterization on the second generation inhibitor (+)-1 (L-, IC50=0.4 nM) and its enantiomer (-)-1 (L-, IC50=43 nM) and their cocrystal structures with PDE4D at 2.0 A resolution. Despite the 107-fold affinity difference, both enantiomers interact with the same sets of residues in the rigid active site. The weaker (-)-1 adopts an unfavorable conformation to preserve the pivotal interactions between the Mg-bound waters and the N-oxide of pyridine. These structures support a model in which inhibitors are anchored by the invariant glutamine at one end and the metal-pocket residues at another end. This model provides explanations for most of the observed structure-activity relationship and the metal ion dependency of the catechol-ether based inhibitors and should facilitate their further design.
Figures


References
-
- Soderling SH, Beavo JA. Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr Opin Cell Biol. 2000;12:174–179. - PubMed
-
- Conti M, Richter W, Mehats C, Livera G, Park JY, Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J Biol Chem. 2003;278:5493–5496. - PubMed
-
- Maurice DH, Palmer D, Tilley DG, Dunkerley HA, Netherton SJ, Raymond DR, Elbatarny HS, Jimmo SL. Cyclic nucleotide phosphodiesterase activity, expression, and targeting in cells of the cardiovascular system. Mol Pharmacol. 2003;64:533–546. - PubMed
-
- Goraya TA, Cooper DM. Ca2+-calmodulin-dependent phosphodiesterase (PDE1): current perspectives. Cell Signal. 2005;17:789–797. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Molecular Biology Databases

