Fast "coalescent" simulation
- PMID: 16539698
- PMCID: PMC1458357
- DOI: 10.1186/1471-2156-7-16
Fast "coalescent" simulation
Abstract
Background: The amount of genome-wide molecular data is increasing rapidly, as is interest in developing methods appropriate for such data. There is a consequent increasing need for methods that are able to efficiently simulate such data. In this paper we implement the sequentially Markovian coalescent algorithm described by McVean and Cardin and present a further modification to that algorithm which slightly improves the closeness of the approximation to the full coalescent model. The algorithm ignores a class of recombination events known to affect the behavior of the genealogy of the sample, but which do not appear to affect the behavior of generated samples to any substantial degree.
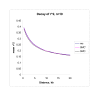
Results: We show that our software is able to simulate large chromosomal regions, such as those appropriate in a consideration of genome-wide data, in a way that is several orders of magnitude faster than existing coalescent algorithms.
Conclusion: This algorithm provides a useful resource for those needing to simulate large quantities of data for chromosomal-length regions using an approach that is much more efficient than traditional coalescent models.
Figures
References
-
- Kingman JFC. On the genealogy of large populations. J Appl Prob. 1982;19A:27–43.
-
- Jiang R, Marjoram P, Stram D. "New data from old" – simulation of test data for mapping studies. 2005.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

