Exploring supervised and unsupervised methods to detect topics in biomedical text
- PMID: 16539745
- PMCID: PMC1472693
- DOI: 10.1186/1471-2105-7-140
Exploring supervised and unsupervised methods to detect topics in biomedical text
Abstract
Background: Topic detection is a task that automatically identifies topics (e.g., "biochemistry" and "protein structure") in scientific articles based on information content. Topic detection will benefit many other natural language processing tasks including information retrieval, text summarization and question answering; and is a necessary step towards the building of an information system that provides an efficient way for biologists to seek information from an ocean of literature.
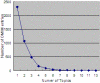
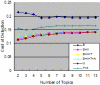
Results: We have explored the methods of Topic Spotting, a task of text categorization that applies the supervised machine-learning technique naïve Bayes to assign automatically a document into one or more predefined topics; and Topic Clustering, which apply unsupervised hierarchical clustering algorithms to aggregate documents into clusters such that each cluster represents a topic. We have applied our methods to detect topics of more than fifteen thousand of articles that represent over sixteen thousand entries in the Online Mendelian Inheritance in Man (OMIM) database. We have explored bag of words as the features. Additionally, we have explored semantic features; namely, the Medical Subject Headings (MeSH) that are assigned to the MEDLINE records, and the Unified Medical Language System (UMLS) semantic types that correspond to the MeSH terms, in addition to bag of words, to facilitate the tasks of topic detection. Our results indicate that incorporating the MeSH terms and the UMLS semantic types as additional features enhances the performance of topic detection and the naïve Bayes has the highest accuracy, 66.4%, for predicting the topic of an OMIM article as one of the total twenty-five topics.
Conclusion: Our results indicate that the supervised topic spotting methods outperformed the unsupervised topic clustering; on the other hand, the unsupervised topic clustering methods have the advantages of being robust and applicable in real world settings.
Figures


References
-
- Smink LJ, Helton EM, Healy BC, Cavnor CC, Lam AC, Flamez D, Burren OS, Wang Y, Dolman GE, Burdick DB, Everett VH, Glusman G, Laneri D, Rowen L, Schuilenburg H, Walker NM, Mychaleckyj J, Wicker LS, Eizirik DL, Todd JA, Goodman N. T1DBase, a community web-based resource for type 1 diabetes research. Nucleic Acids Res. 2005;33:D544–9. doi: 10.1093/nar/gki095. - DOI - PMC - PubMed
-
- Yu H, Hatzivassiloglou V. Towards answering opinion questions: Separating facts from opinions and identifying the polarity of opinion sentences. 2003.
-
- Joachims T. Text categorization with support vector machines: Learning with many relevant features. 1998. pp. 137–142.
-
- Wilbur WJ. A thematic analysis of the AIDS literature. Pac Symp Biocomput. 2002:386–397. - PubMed
-
- Hearst M. The BioText project. A powerpoint presentation.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

