Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists
- PMID: 16540562
- PMCID: PMC6673968
- DOI: 10.1523/JNEUROSCI.0123-06.2005
Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists
Abstract
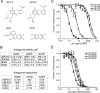
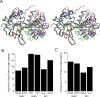
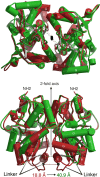
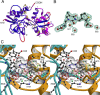
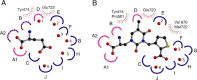
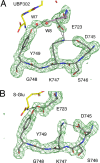
Glutamate receptor (GluR) ion channels mediate fast synaptic transmission in the mammalian CNS. Numerous crystallographic studies, the majority on the GluR2-subtype AMPA receptor, have revealed the structural basis for binding of subtype-specific agonists. In contrast, because there are far fewer antagonist-bound structures, the mechanisms for antagonist binding are much less well understood, particularly for kainate receptors that exist as multiple subtypes with a distinct biology encoded by the GluR5-7, KA1, and KA2 genes. We describe here high-resolution crystal structures for the GluR5 ligand-binding core complex with UBP302 and UBP310, novel GluR5-selective antagonists. The crystal structures reveal the structural basis for the high selectivity for GluR5 observed in radiolabel displacement assays for the isolated ligand binding cores of the GluR2, GluR5, and GluR6 subunits and during inhibition of glutamate-activated currents in studies on full-length ion channels. The antagonists bind via a novel mechanism and do not form direct contacts with the E723 side chain as occurs in all previously solved AMPA and kainate receptor agonist and antagonist complexes. This results from a hyperextension of the ligand binding core compared with previously solved structures. As a result, in dimer assemblies, there is a 22 A extension of the ion channel linkers in the transition from antagonist- to glutamate-bound forms. This large conformational change is substantially different from that described for AMPA receptors, was not possible to predict from previous work, and suggests that glutamate receptors are capable of much larger movements than previously thought.
Figures

Similar articles
-
Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity.Neuron. 2005 Feb 17;45(4):539-52. doi: 10.1016/j.neuron.2005.01.031. Neuron. 2005. PMID: 15721240
-
Tetrazolyl isoxazole amino acids as ionotropic glutamate receptor antagonists: synthesis, modelling and molecular pharmacology.Bioorg Med Chem. 2005 Sep 15;13(18):5391-8. doi: 10.1016/j.bmc.2005.06.024. Bioorg Med Chem. 2005. PMID: 16043357
-
Structure and assembly mechanism for heteromeric kainate receptors.Neuron. 2011 Jul 28;71(2):319-31. doi: 10.1016/j.neuron.2011.05.038. Neuron. 2011. PMID: 21791290 Free PMC article.
-
New developments in the molecular pharmacology of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate and kainate receptors.Pharmacol Ther. 1996;70(1):65-89. doi: 10.1016/0163-7258(96)00014-9. Pharmacol Ther. 1996. PMID: 8804111 Review.
-
The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors.Mol Neurobiol. 2010 Dec;42(3):161-84. doi: 10.1007/s12035-010-8149-x. Epub 2010 Nov 16. Mol Neurobiol. 2010. PMID: 21080238 Free PMC article. Review.
Cited by
-
Emerging structural insights into the function of ionotropic glutamate receptors.Trends Biochem Sci. 2015 Jun;40(6):328-37. doi: 10.1016/j.tibs.2015.04.002. Epub 2015 May 1. Trends Biochem Sci. 2015. PMID: 25941168 Free PMC article. Review.
-
Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics.Structure. 2013 Oct 8;21(10):1788-99. doi: 10.1016/j.str.2013.07.011. Epub 2013 Aug 22. Structure. 2013. PMID: 23972471 Free PMC article.
-
In silico insights of L-glutamate: structural features in vacuum and in complex with its receptor.J Amino Acids. 2013;2013:872058. doi: 10.1155/2013/872058. Epub 2013 Nov 6. J Amino Acids. 2013. PMID: 24307941 Free PMC article.
-
Distinct receptors underlie glutamatergic signalling in inspiratory rhythm-generating networks and motor output pathways in neonatal rat.J Physiol. 2008 May 1;586(9):2357-70. doi: 10.1113/jphysiol.2007.150532. Epub 2008 Mar 13. J Physiol. 2008. PMID: 18339693 Free PMC article.
-
Molecular basis of kainate receptor modulation by sodium.Neuron. 2008 Jun 12;58(5):720-35. doi: 10.1016/j.neuron.2008.04.001. Neuron. 2008. PMID: 18549784 Free PMC article.
References
-
- Anslyn EV, Dougherty DA (2005). In: Modern physical organic chemistry Sausalito, CA: University Science Books.
-
- Arinaminpathy Y, Sansom MS, Biggin PC (2006). Binding site flexibility: molecular simulation of partial and full agonists with a glutamate receptor. Mol Pharmacol 69:5–12. - PubMed
-
- Armstrong N, Gouaux E (2000). Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28:165–181. - PubMed
-
- Brauner-Osborne H, Egebjerg J, Nielsen EO, Madsen U, Krogsgaard-Larsen P (2000). Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem 43:2609–2645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
