Control of mammalian translation by mRNA structure near caps
- PMID: 16540693
- PMCID: PMC1440912
- DOI: 10.1261/rna.2309906
Control of mammalian translation by mRNA structure near caps
Abstract
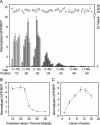
The scanning model of RNA translation proposes that highly stable secondary structures within mRNAs can inhibit translation, while structures of lower thermal stability also affect translation if close enough to the 5' methyl G cap. However, only fragmentary information is available about the dependence of translation efficiency in live mammalian cells on the thermodynamic stability, location, and GC content of RNA structures in the 5'-untranslated region. We devised a two-color fluorescence assay for translation efficiency in single live cells and compared a wide range of hairpins with predicted thermal stabilities ranging from -10 to -50 kcal/mol and 5' G cap-to-hairpin distances of 1-46 bases. Translation efficiency decreased abruptly as hairpin stabilities increased from deltaG = -25 to -35 kcal/mol. Shifting a hairpin as little as nine bases relative to the 5' cap could modulate translation more than 50-fold. Increasing GC content diminished translation efficiency when predicted thermal stability and cap-to-hairpin distances were held constant. We additionally found naturally occurring 5'-untranslated regions affected translation differently in live cells compared with translation in in vitro lysates. Our study will assist scientists in designing experiments that deliberately modulate mammalian translation with designed 5' UTRs.
Figures



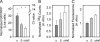


References
-
- Bayer T.S., Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. - PubMed
-
- Brantl S. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta. 2002;1575:15–25. - PubMed
-
- Davidson E.A., Ellington A.D. Engineering regulatory RNAs. Trends Biotechnol. 2005;23:109–112. - PubMed
-
- de Benedetti A., Harris A.L. eIF4E expression in tumors: Its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 1999;31:59–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
