A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA)
- PMID: 16540696
- PMCID: PMC1440900
- DOI: 10.1261/rna.2332406
A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA)
Abstract
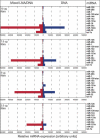
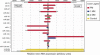
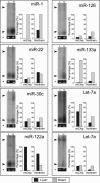
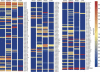
MicroRNAs represent a class of short (approximately 22 nt), noncoding regulatory RNAs involved in development, differentiation, and metabolism. We describe a novel microarray platform for genome-wide profiling of mature miRNAs (miChip) using locked nucleic acid (LNA)-modified capture probes. The biophysical properties of LNA were exploited to design probe sets for uniform, high-affinity hybridizations yielding highly accurate signals able to discriminate between single nucleotide differences and, hence, between closely related miRNA family members. The superior detection sensitivity eliminates the need for RNA size selection and/or amplification. MiChip will greatly simplify miRNA expression profiling of biological and clinical samples.
Figures
References
-
- Braasch D.A., Corey D.R. Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
