INTEGRATOR: interactive graphical search of large protein interactomes over the Web
- PMID: 16542421
- PMCID: PMC1459205
- DOI: 10.1186/1471-2105-7-146
INTEGRATOR: interactive graphical search of large protein interactomes over the Web
Abstract
Background: The rapid growth of protein interactome data has elevated the necessity and importance of network analysis tools. However, unlike pure text data, network search spaces are of exponential complexity. This poses special challenges for storing, searching, and navigating this data efficiently. Moreover, development of effective web interfaces has been difficult.
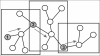
Results: We present Integrator, a web-integrated graphical search tool for protein-protein interaction networks across 50+ genomes.
Conclusion: Integrator provides single and multiple protein searches of the Bioverse database containing experimentally-derived and predicted protein-protein interactions. The interface provides animated local network views, rapid subgraph manipulation, and cross-referencing of functional annotations. Integrator is available at http://bioverse.compbio.washington.edu/integrator.
Figures



Similar articles
-
MannDB - a microbial database of automated protein sequence analyses and evidence integration for protein characterization.BMC Bioinformatics. 2006 Oct 17;7:459. doi: 10.1186/1471-2105-7-459. BMC Bioinformatics. 2006. PMID: 17044936 Free PMC article.
-
SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces.BMC Bioinformatics. 2006 Mar 2;7:104. doi: 10.1186/1471-2105-7-104. BMC Bioinformatics. 2006. PMID: 16512892 Free PMC article.
-
BIOVERSE: enhancements to the framework for structural, functional and contextual modeling of proteins and proteomes.Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W324-5. doi: 10.1093/nar/gki401. Nucleic Acids Res. 2005. PMID: 15980482 Free PMC article.
-
Biomolecular interaction network database.Brief Bioinform. 2005 Jun;6(2):194-8. doi: 10.1093/bib/6.2.194. Brief Bioinform. 2005. PMID: 15975228 Review.
-
Searching the MINT database for protein interaction information.Curr Protoc Bioinformatics. 2008 Jun;Chapter 8:8.5.1-8.5.13. doi: 10.1002/0471250953.bi0805s22. Curr Protoc Bioinformatics. 2008. PMID: 18551417 Review.
Cited by
-
SynechoNET: integrated protein-protein interaction database of a model cyanobacterium Synechocystis sp. PCC 6803.BMC Bioinformatics. 2008;9 Suppl 1(Suppl 1):S20. doi: 10.1186/1471-2105-9-S1-S20. BMC Bioinformatics. 2008. PMID: 18315852 Free PMC article.
-
Network integration and graph analysis in mammalian molecular systems biology.IET Syst Biol. 2008 Sep;2(5):206-21. doi: 10.1049/iet-syb:20070075. IET Syst Biol. 2008. PMID: 19045817 Free PMC article. Review.
References
-
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

