Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex
- PMID: 16547172
- PMCID: PMC1475731
- DOI: 10.1101/gad.1399406
Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex
Abstract
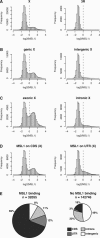
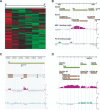
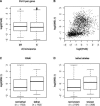
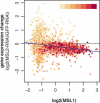
The dosage compensation complex (DCC) of Drosophila melanogaster is capable of distinguishing the single male X from the other chromosomes in the nucleus. It selectively interacts in a discontinuous pattern with much of the X chromosome. How the DCC identifies and binds the X, including binding to the many genes that require dosage compensation, is currently unknown. To identify bound genes and attempt to isolate the targeting cues, we visualized male-specific lethal 1 (MSL1) protein binding along the X chromosome by combining chromatin immunoprecipitation with high-resolution microarrays. More than 700 binding regions for the DCC were observed, encompassing more than half the genes found on the X chromosome. In addition, several rare autosomal binding sites were identified. Essential genes are preferred targets, and genes binding high levels of DCC appear to experience the most compensation (i.e., greatest increase in expression). DCC binding clearly favors genes over intergenic regions, and binds most strongly to the 3' end of transcription units. Within the targeted genes, the DCC exhibits a strong preference for exons and coding sequences. Our results demonstrate gene-specific binding of the DCC, and identify several sequence elements that may partly direct its targeting.
Figures



References
-
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Akhtar A., Becker P.B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell. 2000;5:367–375. - PubMed
-
- Arbeitman M.N., Furlong E.E., Imam F., Johnson E., Null B.H., Baker B.S., Krasnow M.A., Scott M.P., Davis R.W., White K.P. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. - PubMed
-
- Baker B.S., Gorman M., Marin I. Dosage compensation in Drosophila. Annu. Rev. Genet. 1994;28:491–521. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
