High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome
- PMID: 16547173
- PMCID: PMC1472287
- DOI: 10.1101/gad.1400206
High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome
Abstract
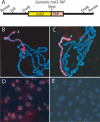
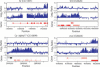
X-chromosome dosage compensation in Drosophila requires the male-specific lethal (MSL) complex, which up-regulates gene expression from the single male X chromosome. Here, we define X-chromosome-specific MSL binding at high resolution in two male cell lines and in late-stage embryos. We find that the MSL complex is highly enriched over most expressed genes, with binding biased toward the 3' end of transcription units. The binding patterns are largely similar in the distinct cell types, with approximately 600 genes clearly bound in all three cases. Genes identified as clearly bound in one cell type and not in another indicate that attraction of MSL complex correlates with expression state. Thus, sequence alone is not sufficient to explain MSL targeting. We propose that the MSL complex recognizes most X-linked genes, but only in the context of chromatin factors or modifications indicative of active transcription. Distinguishing expressed genes from the bulk of the genome is likely to be an important function common to many chromatin organizing and modifying activities.
Figures




References
-
- Bone J.R., Lavender J., Richman R., Palmer M.J., Turner B.M., Kuroda M.I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes & Dev. 1994;8:96–104. - PubMed
-
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. - PubMed
-
- Currie D., Milner M., Evans C. The growth and differentiation in vitro of leg and wing imaginal disc cells from Drosophila melanogaster. Development. 1988;102:805–814.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
