An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis
- PMID: 16547197
- PMCID: PMC1409679
- DOI: 10.1093/nar/gkl050
An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis
Abstract
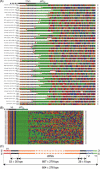
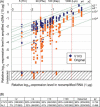
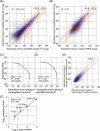
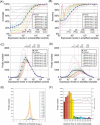
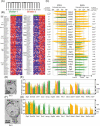
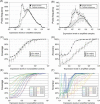
A systems-level understanding of a small but essential population of cells in development or adulthood (e.g. somatic stem cells) requires accurate quantitative monitoring of genome-wide gene expression, ideally from single cells. We report here a strategy to globally amplify mRNAs from single cells for highly quantitative high-density oligonucleotide microarray analysis that combines a small number of directional PCR cycles with subsequent linear amplification. Using this strategy, both the representation of gene expression profiles and reproducibility between individual experiments are unambiguously improved from the original method, along with high coverage and accuracy. The immediate application of this method to single cells in the undifferentiated inner cell masses of mouse blastocysts at embryonic day (E) 3.5 revealed the presence of two populations of cells, one with primitive endoderm (PE) expression and the other with pluripotent epiblast-like gene expression. The genes expressed differentially between these two populations were well preserved in morphologically differentiated PE and epiblast in the embryos one day later (E4.5), demonstrating that the method successfully detects subtle but essential differences in gene expression at the single-cell level among seemingly homogeneous cell populations. This study provides a strategy to analyze biophysical events in medicine as well as in neural, stem cell and developmental biology, where small numbers of distinctive or diseased cells play critical roles.
Figures

References
-
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. - PubMed
-
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. - PubMed
-
- Westerhoff H.V., Palsson B.O. The evolution of molecular biology into systems biology. Nat. Biotechnol. 2004;22:1249–1252. - PubMed
-
- Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

