MicroRNA fingerprints during human megakaryocytopoiesis
- PMID: 16549775
- PMCID: PMC1458797
- DOI: 10.1073/pnas.0600587103
MicroRNA fingerprints during human megakaryocytopoiesis
Abstract
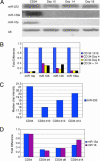
microRNAs are a highly conserved class of noncoding RNAs with important regulatory functions in proliferation, apoptosis, development, and differentiation. To discover novel regulatory pathways during megakaryocytic differentiation, we performed microRNA expression profiling of in vitro-differentiated megakaryocytes derived from CD34(+) hematopoietic progenitors. The main finding was down-regulation of miR-10a, miR-126, miR-106, miR-10b, miR-17 and miR-20. Hypothetically, the down-regulation of microRNAs unblocks target genes involved in differentiation. We confirmed in vitro and in vivo that miR-130a targets the transcription factor MAFB, which is involved in the activation of the GPIIB promoter, a key protein for platelet physiology. In addition, we found that miR-10a expression in differentiated megakaryocytes is inverse to that of HOXA1, and we showed that HOXA1 is a direct target of miR-10a. Finally, we compared the microRNA expression of megakaryoblastic leukemic cell lines with that of in vitro differentiated megakaryocytes and CD34(+) progenitors. This analysis revealed up-regulation of miR-101, miR-126, miR-99a, miR-135, and miR-20. Our data delineate the expression of microRNAs during megakaryocytopoiesis and suggest a regulatory role of microRNAs in this process by targeting megakaryocytic transcription factors.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

