In vitro selection of tRNAs for efficient four-base decoding to incorporate non-natural amino acids into proteins in an Escherichia coli cell-free translation system
- PMID: 16549877
- PMCID: PMC1405820
- DOI: 10.1093/nar/gkl087
In vitro selection of tRNAs for efficient four-base decoding to incorporate non-natural amino acids into proteins in an Escherichia coli cell-free translation system
Abstract
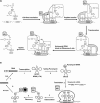
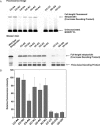
Position-specific incorporation of non-natural amino acids into proteins is a useful technique in protein engineering. In this study, we established a novel selection system to obtain tRNAs that show high decoding activity, from a tRNA library in a cell-free translation system to improve the efficiency of incorporation of non-natural amino acids into proteins. In this system, a puromycin-tRNA conjugate, in which the 3'-terminal A unit was replaced by puromycin, was used. The puromycin-tRNA conjugate was fused to a C-terminus of streptavidin through the puromycin moiety in the ribosome. The streptavidin-puromycin-tRNA fusion molecule was collected and brought to the next round after amplification of the tRNA sequence. We applied this system to select efficient frameshift suppressor tRNAs from a tRNA library with a randomly mutated anticodon loop derived from yeast tRNA CCCG Phe. After three rounds of the selection, we obtained novel frameshift suppressor tRNAs which had high decoding activity and good orthogonality against endogenous aminoacyl-tRNA synthetases. These results demonstrate that the in vitro selection system developed here is useful to obtain highly active tRNAs for the incorporation of non-natural amino acid from a tRNA library.
Figures



Similar articles
-
Comprehensive screening of amber suppressor tRNAs suitable for incorporation of non-natural amino acids in a cell-free translation system.Biochem Biophys Res Commun. 2008 Sep 19;374(2):304-8. doi: 10.1016/j.bbrc.2008.07.020. Epub 2008 Jul 15. Biochem Biophys Res Commun. 2008. PMID: 18634752
-
Four-base codon-mediated saturation mutagenesis in a cell-free translation system.J Biosci Bioeng. 2008 Mar;105(3):211-5. doi: 10.1263/jbb.105.211. J Biosci Bioeng. 2008. PMID: 18397770
-
Multiple incorporation of non-natural amino acids into a single protein using tRNAs with non-standard structures.FEBS Lett. 2005 Dec 19;579(30):6769-74. doi: 10.1016/j.febslet.2005.11.010. Epub 2005 Nov 21. FEBS Lett. 2005. PMID: 16310775
-
tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations.Biochimie. 2005 Sep-Oct;87(9-10):835-45. doi: 10.1016/j.biochi.2005.02.014. Biochimie. 2005. PMID: 15925436 Review.
-
tRNA's wobble decoding of the genome: 40 years of modification.J Mol Biol. 2007 Feb 9;366(1):1-13. doi: 10.1016/j.jmb.2006.11.046. Epub 2006 Nov 15. J Mol Biol. 2007. PMID: 17187822 Review.
Cited by
-
Cellular Site-Specific Incorporation of Noncanonical Amino Acids in Synthetic Biology.Chem Rev. 2024 Sep 25;124(18):10577-10617. doi: 10.1021/acs.chemrev.3c00938. Epub 2024 Aug 29. Chem Rev. 2024. PMID: 39207844 Review.
-
Genetic Code Expansion Through Quadruplet Codon Decoding.J Mol Biol. 2022 Apr 30;434(8):167346. doi: 10.1016/j.jmb.2021.167346. Epub 2021 Nov 8. J Mol Biol. 2022. PMID: 34762896 Free PMC article. Review.
-
A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment.Microbiol Mol Biol Rev. 2009 Mar;73(1):178-210. doi: 10.1128/MMBR.00010-08. Microbiol Mol Biol Rev. 2009. PMID: 19258537 Free PMC article. Review.
-
A Concept for Selection of Codon-Suppressor tRNAs Based on Read-Through Ribosome Display in an In Vitro Compartmentalized Cell-Free Translation System.J Nucleic Acids. 2012;2012:538129. doi: 10.1155/2012/538129. Epub 2012 Jul 18. J Nucleic Acids. 2012. PMID: 22928090 Free PMC article.
-
Genetically encoded libraries of nonstandard peptides.J Nucleic Acids. 2012;2012:713510. doi: 10.1155/2012/713510. Epub 2012 Oct 14. J Nucleic Acids. 2012. PMID: 23097693 Free PMC article.
References
-
- Cornish V.W., Mendel D., Schultz P.G. Probing protein structure and function with an expanded genetic code. Angew. Chem., Int. Ed. Engl. 1995;34:621–633.
-
- Steward L.E., Chamberlin A.R. Protein engineering with nonstandard amino acids. Methods Mol. Biol. 1998;77:325–354. - PubMed
-
- Anderson R.D., III, Zhou J., Hecht S.M. Fluorescence resonance energy transfer between unnatural amino acids in a structurally modified dihydrofolate reductase. J. Am. Chem. Soc. 2002;124:9674–9675. - PubMed
-
- Hohsaka T., Muranaka N., Komiyama C., Matsui K., Takaura S., Abe R., Murakami H., Sisido M. Position-specific incorporation of dansylated non-natural amino acids into streptavidin by using a four-base codon. FEBS Lett. 2004;560:173–177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

