Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays
- PMID: 16551369
- PMCID: PMC1435885
- DOI: 10.1186/1471-2164-7-59
Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays
Abstract
Background: DNA microarrays are rapidly becoming a fundamental tool in discovery-based genomic and biomedical research. However, the reliability of the microarray results is being challenged due to the existence of different technologies and non-standard methods of data analysis and interpretation. In the absence of a "gold standard"/"reference method" for the gene expression measurements, studies evaluating and comparing the performance of various microarray platforms have often yielded subjective and conflicting conclusions. To address this issue we have conducted a large scale TaqMan Gene Expression Assay based real-time PCR experiment and used this data set as the reference to evaluate the performance of two representative commercial microarray platforms.
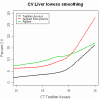
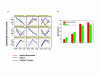
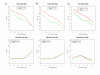
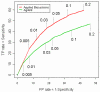
Results: In this study, we analyzed the gene expression profiles of three human tissues: brain, lung, liver and one universal human reference sample (UHR) using two representative commercial long-oligonucleotide microarray platforms: (1) Applied Biosystems Human Genome Survey Microarrays (based on single-color detection); (2) Agilent Whole Human Genome Oligo Microarrays (based on two-color detection). 1,375 genes represented by both microarray platforms and spanning a wide dynamic range in gene expression levels, were selected for TaqMan Gene Expression Assay based real-time PCR validation. For each platform, four technical replicates were performed on the same total RNA samples according to each manufacturer's standard protocols. For Agilent arrays, comparative hybridization was performed using incorporation of Cy5 for brain/lung/liver RNA and Cy3 for UHR RNA (common reference). Using the TaqMan Gene Expression Assay based real-time PCR data set as the reference set, the performance of the two microarray platforms was evaluated focusing on the following criteria: (1) Sensitivity and accuracy in detection of expression; (2) Fold change correlation with real-time PCR data in pair-wise tissues as well as in gene expression profiles determined across all tissues; (3) Sensitivity and accuracy in detection of differential expression.
Conclusion: Our study provides one of the largest "reference" data set of gene expression measurements using TaqMan Gene Expression Assay based real-time PCR technology. This data set allowed us to use an alternative gene expression technology to evaluate the performance of different microarray platforms. We conclude that microarrays are indeed invaluable discovery tools with acceptable reliability for genome-wide gene expression screening, though validation of putative changes in gene expression remains advisable. Our study also characterizes the limitations of microarrays; understanding these limitations will enable researchers to more effectively evaluate microarray results in a more cautious and appropriate manner.
Figures




References
-
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino G, Griffin C, Hilmer SC, Hoffman E, Jedlicka AE, Kawasaki E, Martinez-Murillo F, Morsberger L, Lee H, Petersen D, Quackenbush J, Scott A, Wilson M, Yang Y, Ye SQ, Yu W. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

