SNPs3D: candidate gene and SNP selection for association studies
- PMID: 16551372
- PMCID: PMC1435944
- DOI: 10.1186/1471-2105-7-166
SNPs3D: candidate gene and SNP selection for association studies
Abstract
Background: The relationship between disease susceptibility and genetic variation is complex, and many different types of data are relevant. We describe a web resource and database that provides and integrates as much information as possible on disease/gene relationships at the molecular level.
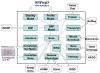
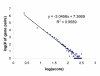
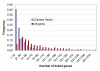
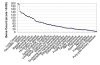
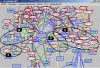
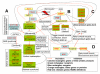
Description: The resource http://www.SNPs3D.org has three primary modules. One module identifies which genes are candidates for involvement in a specified disease. A second module provides information about the relationships between sets of candidate genes. The third module analyzes the likely impact of non-synonymous SNPs on protein function. Disease/candidate gene relationships and gene-gene relationships are derived from the literature using simple but effective text profiling. SNP/protein function relationships are derived by two methods, one using principles of protein structure and stability, the other based on sequence conservation. Entries for each gene include a number of links to other data, such as expression profiles, pathway context, mouse knockout information and papers. Gene-gene interactions are presented in an interactive graphical interface, providing rapid access to the underlying information, as well as convenient navigation through the network. Use of the resource is illustrated with aspects of the inflammatory response and hypertension.
Conclusion: The combination of SNP impact analysis, a knowledge based network of gene relationships and candidate genes, and access to a wide range of data and literature allow a user to quickly assimilate available information, and so develop models of gene-pathway-disease interaction.
Figures

Similar articles
-
LDGIdb: a database of gene interactions inferred from long-range strong linkage disequilibrium between pairs of SNPs.BMC Res Notes. 2012 May 2;5:212. doi: 10.1186/1756-0500-5-212. BMC Res Notes. 2012. PMID: 22551073 Free PMC article.
-
An integrated database-pipeline system for studying single nucleotide polymorphisms and diseases.BMC Bioinformatics. 2008 Dec 12;9 Suppl 12(Suppl 12):S19. doi: 10.1186/1471-2105-9-S12-S19. BMC Bioinformatics. 2008. PMID: 19091018 Free PMC article.
-
SUSPECTS: enabling fast and effective prioritization of positional candidates.Bioinformatics. 2006 Mar 15;22(6):773-4. doi: 10.1093/bioinformatics/btk031. Epub 2006 Jan 19. Bioinformatics. 2006. PMID: 16423925
-
MutaGeneSys: estimating individual disease susceptibility based on genome-wide SNP array data.Bioinformatics. 2008 Feb 1;24(3):440-2. doi: 10.1093/bioinformatics/btm587. Epub 2007 Nov 29. Bioinformatics. 2008. PMID: 18048395
-
[Genetic aspects of erectile dysfunction].Urologe A. 2015 May;54(5):662-7. doi: 10.1007/s00120-015-3793-4. Urologe A. 2015. PMID: 25987331 Review. German.
Cited by
-
Predicting folding free energy changes upon single point mutations.Bioinformatics. 2012 Mar 1;28(5):664-71. doi: 10.1093/bioinformatics/bts005. Epub 2012 Jan 11. Bioinformatics. 2012. PMID: 22238268 Free PMC article.
-
Candidate gene association studies: a comprehensive guide to useful in silico tools.BMC Genet. 2013 May 9;14:39. doi: 10.1186/1471-2156-14-39. BMC Genet. 2013. PMID: 23656885 Free PMC article. Review.
-
Investigating the structural impacts of I64T and P311S mutations in APE1-DNA complex: a molecular dynamics approach.PLoS One. 2012;7(2):e31677. doi: 10.1371/journal.pone.0031677. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22384055 Free PMC article.
-
The role of balanced training and testing data sets for binary classifiers in bioinformatics.PLoS One. 2013 Jul 9;8(7):e67863. doi: 10.1371/journal.pone.0067863. Print 2013. PLoS One. 2013. PMID: 23874456 Free PMC article.
-
Towards Increasing the Clinical Relevance of In Silico Methods to Predict Pathogenic Missense Variants.PLoS Comput Biol. 2016 May 12;12(5):e1004725. doi: 10.1371/journal.pcbi.1004725. eCollection 2016 May. PLoS Comput Biol. 2016. PMID: 27171182 Free PMC article. Review. No abstract available.
References
-
- SNPs3D. [http:/wwwsnps3dorg]
-
- Biomolecular Interaction Network Database (BIND) [http://bindca]
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

