Prediction of indirect interactions in proteins
- PMID: 16553946
- PMCID: PMC1435945
- DOI: 10.1186/1471-2105-7-167
Prediction of indirect interactions in proteins
Abstract
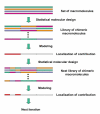
Background: Both direct and indirect interactions determine molecular recognition of ligands by proteins. Indirect interactions can be defined as effects on recognition controlled from distant sites in the proteins, e.g. by changes in protein conformation and mobility, whereas direct interactions occur in close proximity of the protein's amino acids and the ligand. Molecular recognition is traditionally studied using three-dimensional methods, but with such techniques it is difficult to predict the effects caused by mutational changes of amino acids located far away from the ligand-binding site. We recently developed an approach, proteochemometrics, to the study of molecular recognition that models the chemical effects involved in the recognition of ligands by proteins using statistical sampling and mathematical modelling.
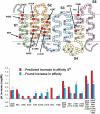
Results: A proteochemometric model was built, based on a statistically designed protein library's (melanocortin receptors') interaction with three peptides and used to predict which amino acids and sequence fragments that are involved in direct and indirect ligand interactions. The model predictions were confirmed by directed mutagenesis. The predicted presumed direct interactions were in good agreement with previous three-dimensional studies of ligand recognition. However, in addition the model could also correctly predict the location of indirect effects on ligand recognition arising from distant sites in the receptors, something that three-dimensional modelling could not afford.
Conclusion: We demonstrate experimentally that proteochemometric modelling can be used with high accuracy to predict the site of origin of direct and indirect effects on ligand recognitions by proteins.
Figures


References
-
- Prusis P, Muceniece R, Andersson P, Post C, Lundstedt T, Wikberg JE. PLS modeling of chimeric MS04/MSH-peptide and MC1/MC3-receptor interactions reveals a novel method for the analysis of ligand-receptor interactions. Biochim Biophys Acta. 2001;1544:350–357. - PubMed
-
- Wikberg JES, Lapinsh M, Prusis P. Proteochemometrics: A tool for modelling the molecular interaction space. In: Kubinyi H, Müller G, Mannhold R, Folkers G, editor. Chemogenomics in Drug Discovery – A Medicinal Chemistry Perspective. Weinheim: Wiley-VCH; 2004. pp. 289–309. - PubMed
-
- Lapinsh M, Prusis P, Gutcaits A, Lundstedt T, Wikberg JE. Development of proteo-chemometrics: a novel technology for the analysis of drug-receptor interactions. Biochim Biophys Acta. 2001;1525:180–190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

