Origin and evolution of the peroxisomal proteome
- PMID: 16556314
- PMCID: PMC1472686
- DOI: 10.1186/1745-6150-1-8
Origin and evolution of the peroxisomal proteome
Abstract
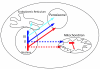
Background: Peroxisomes are ubiquitous eukaryotic organelles involved in various oxidative reactions. Their enzymatic content varies between species, but the presence of common protein import and organelle biogenesis systems support a single evolutionary origin. The precise scenario for this origin remains however to be established. The ability of peroxisomes to divide and import proteins post-translationally, just like mitochondria and chloroplasts, supports an endosymbiotic origin. However, this view has been challenged by recent discoveries that mutant, peroxisome-less cells restore peroxisomes upon introduction of the wild-type gene, and that peroxisomes are formed from the Endoplasmic Reticulum. The lack of a peroxisomal genome precludes the use of classical analyses, as those performed with mitochondria or chloroplasts, to settle the debate. We therefore conducted large-scale phylogenetic analyses of the yeast and rat peroxisomal proteomes.
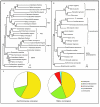
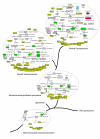
Results: Our results show that most peroxisomal proteins (39-58%) are of eukaryotic origin, comprising all proteins involved in organelle biogenesis or maintenance. A significant fraction (13-18%), consisting mainly of enzymes, has an alpha-proteobacterial origin and appears to be the result of the recruitment of proteins originally targeted to mitochondria. Consistent with the findings that peroxisomes are formed in the Endoplasmic Reticulum, we find that the most universally conserved Peroxisome biogenesis and maintenance proteins are homologous to proteins from the Endoplasmic Reticulum Assisted Decay pathway.
Conclusion: Altogether our results indicate that the peroxisome does not have an endosymbiotic origin and that its proteins were recruited from pools existing within the primitive eukaryote. Moreover the reconstruction of primitive peroxisomal proteomes suggests that ontogenetically as well as phylogenetically, peroxisomes stem from the Endoplasmic Reticulum.
Reviewers: This article was reviewed by Arcady Mushegian, Gáspár Jékely and John Logsdon.
Open peer review: Reviewed by Arcady Mushegian, Gáspar Jékely and John Logsdon. For the full reviews, please go to the Reviewers' comments section.
Figures

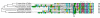
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

