Protein assembly and DNA looping by the FokI restriction endonuclease
- PMID: 16556912
- PMCID: PMC1410913
- DOI: 10.1093/nar/gkl076
Protein assembly and DNA looping by the FokI restriction endonuclease
Abstract
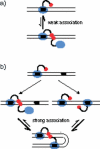
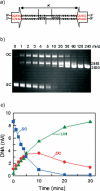
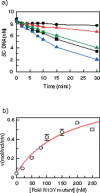
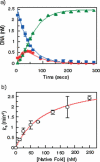
The FokI restriction endonuclease recognizes an asymmetric DNA sequence and cuts both strands at fixed positions upstream of the site. The sequence is contacted by a single monomer of the protein, but the monomer has only one catalytic centre and forms a dimer to cut both strands. FokI is also known to cleave DNA with two copies of its site more rapidly than DNA with one copy. To discover how FokI acts at a single site and how it acts at two sites, its reactions were examined on a series of plasmids with either one recognition site or with two sites separated by varied distances, sometimes in the presence of a DNA-binding defective mutant of FokI. These experiments showed that, to cleave DNA with one site, the monomer bound to that site associates via a weak protein-protein interaction with a second monomer that remains detached from the recognition sequence. Nevertheless, the second monomer catalyses phosphodiester bond hydrolysis at the same rate as the DNA-bound monomer. On DNA with two sites, two monomers of FokI interact strongly, as a result of being tethered to the same molecule of DNA, and sequester the intervening DNA in a loop.
Figures

Similar articles
-
Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands.Nucleic Acids Res. 2009 Apr;37(7):2105-15. doi: 10.1093/nar/gkp046. Epub 2009 Feb 17. Nucleic Acids Res. 2009. PMID: 19223323 Free PMC article.
-
Dynamics and consequences of DNA looping by the FokI restriction endonuclease.Nucleic Acids Res. 2008 Apr;36(6):2073-81. doi: 10.1093/nar/gkn051. Epub 2008 Feb 14. Nucleic Acids Res. 2008. PMID: 18276642 Free PMC article.
-
Illuminating the reaction pathway of the FokI restriction endonuclease by fluorescence resonance energy transfer.Nucleic Acids Res. 2012 Feb;40(3):1203-13. doi: 10.1093/nar/gkr809. Epub 2011 Oct 12. Nucleic Acids Res. 2012. PMID: 21993298 Free PMC article.
-
The reaction mechanism of FokI excludes the possibility of targeting zinc finger nucleases to unique DNA sites.Biochem Soc Trans. 2011 Apr;39(2):584-8. doi: 10.1042/BST0390584. Biochem Soc Trans. 2011. PMID: 21428944 Review.
-
Fluorescent substrates for the EcoRV restriction endonuclease.Biochem Soc Trans. 1994 Aug;22(3):299S. doi: 10.1042/bst022299s. Biochem Soc Trans. 1994. PMID: 7821558 Review. No abstract available.
Cited by
-
Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands.Nucleic Acids Res. 2009 Apr;37(7):2105-15. doi: 10.1093/nar/gkp046. Epub 2009 Feb 17. Nucleic Acids Res. 2009. PMID: 19223323 Free PMC article.
-
TALE-PvuII fusion proteins--novel tools for gene targeting.PLoS One. 2013 Dec 5;8(12):e82539. doi: 10.1371/journal.pone.0082539. eCollection 2013. PLoS One. 2013. PMID: 24349308 Free PMC article.
-
Concerted action at eight phosphodiester bonds by the BcgI restriction endonuclease.Nucleic Acids Res. 2011 Sep 1;39(17):7630-40. doi: 10.1093/nar/gkr453. Epub 2011 Jun 7. Nucleic Acids Res. 2011. PMID: 21653548 Free PMC article.
-
Creating designed zinc-finger nucleases with minimal cytotoxicity.J Mol Biol. 2011 Jan 21;405(3):630-41. doi: 10.1016/j.jmb.2010.10.043. Epub 2010 Nov 19. J Mol Biol. 2011. PMID: 21094162 Free PMC article.
-
Adding fingers to an engineered zinc finger nuclease can reduce activity.Biochemistry. 2011 Jun 7;50(22):5033-41. doi: 10.1021/bi200393g. Epub 2011 May 11. Biochemistry. 2011. PMID: 21528840 Free PMC article.
References
-
- Lagunavicius A., Sasnauskas G., Halford S.E., Siksnys V. The metal-independent Type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. - PubMed
-
- Halford S.E. Hopping, jumping and looping by restriction enzymes. Biochem. Soc. Trans. 2001;29:363–373. - PubMed
-
- Aggarwal A.K. Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol. 1995;5:11–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

