Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins
- PMID: 16563186
- PMCID: PMC1557759
- DOI: 10.1186/gb-2006-7-3-r23
Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins
Abstract
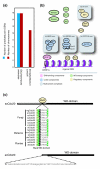
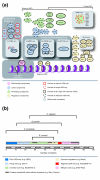
Background: Kinetochores are large multi-protein structures that assemble on centromeric DNA (CEN DNA) and mediate the binding of chromosomes to microtubules. Comprising 125 base-pairs of CEN DNA and 70 or more protein components, Saccharomyces cerevisiae kinetochores are among the best understood. In contrast, most fungal, plant and animal cells assemble kinetochores on CENs that are longer and more complex, raising the question of whether kinetochore architecture has been conserved through evolution, despite considerable divergence in CEN sequence.
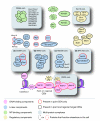
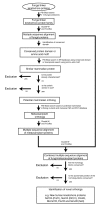
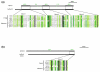
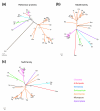
Results: Using computational approaches, ranging from sequence similarity searches to hidden Markov model-based modeling, we show that organisms with CENs resembling those in S. cerevisiae (point CENs) are very closely related and that all contain a set of 11 kinetochore proteins not found in organisms with complex CENs. Conversely, organisms with complex CENs (regional CENs) contain proteins seemingly absent from point-CEN organisms. However, at least three quarters of known kinetochore proteins are present in all fungi regardless of CEN organization. At least six of these proteins have previously unidentified human orthologs. When fungi and metazoa are compared, almost all have kinetochores constructed around Spc105 and three conserved multi-protein linker complexes (MIND, COMA, and the NDC80 complex).
Conclusion: Our data suggest that critical structural features of kinetochores have been well conserved from yeast to man. Surprisingly, phylogenetic analysis reveals that human kinetochore proteins are as similar in sequence to their yeast counterparts as to presumptive Drosophila melanogaster or Caenorhabditis elegans orthologs. This finding is consistent with evidence that kinetochore proteins have evolved very rapidly relative to components of other complex cellular structures.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

