Mutant fibrinogen cleared from the endoplasmic reticulum via endoplasmic reticulum-associated protein degradation and autophagy: an explanation for liver disease
- PMID: 16565503
- PMCID: PMC1606570
- DOI: 10.2353/ajpath.2006.051097
Mutant fibrinogen cleared from the endoplasmic reticulum via endoplasmic reticulum-associated protein degradation and autophagy: an explanation for liver disease
Abstract
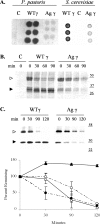
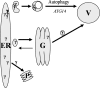
The endoplasmic reticulum (ER) quality control processes recognize and remove aberrant proteins from the secretory pathway. Several variants of the plasma protein fibrinogen are recognized as aberrant and degraded by ER-associated protein degradation (ERAD), thus leading to hypofibrinogenemia. A subset of patients with hypofibrinogenemia exhibit hepatic ER accumulation of the variant fibrinogens and develop liver cirrhosis. One such variant named Aguadilla has a substitution of Arg375 to Trp in the gamma-chain. To understand the cellular mechanisms behind clearance of the aberrant Aguadilla gamma-chain, we expressed the mutant gammaD domain in yeast and found that it was cleared from the ER via ERAD. In addition, we discovered that when ERAD was saturated, aggregated Aguadilla gammaD accumulated within the ER while a soluble form of the polypeptide transited the secretory pathway to the trans-Golgi network where it was targeted to the vacuole for degradation. Examination of Aguadilla gammaD in an autophagy-deficient yeast strain showed stabilization of the aggregated ER form, indicating that these aggregates are normally cleared from the ER via the autophagic pathway. These findings have clinical relevance in the understanding of and treatment for ER storage diseases.
Figures
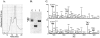



Similar articles
-
Fibrinogen gamma375 arg-->trp mutation (fibrinogen aguadilla) causes hereditary hypofibrinogenemia, hepatic endoplasmic reticulum storage disease and cirrhosis.Am J Surg Pathol. 2006 Jul;30(7):906-11. doi: 10.1097/01.pas.0000209848.59670.2c. Am J Surg Pathol. 2006. PMID: 16819336
-
Novel fibrinogen gamma375 Arg-->Trp mutation (fibrinogen aguadilla) causes hepatic endoplasmic reticulum storage and hypofibrinogenemia.Hepatology. 2002 Sep;36(3):652-8. doi: 10.1053/jhep.2002.35063. Hepatology. 2002. PMID: 12198657
-
Fibrinogen Gamma Chain Mutations Provoke Fibrinogen and Apolipoprotein B Plasma Deficiency and Liver Storage.Int J Mol Sci. 2017 Dec 15;18(12):2717. doi: 10.3390/ijms18122717. Int J Mol Sci. 2017. PMID: 29244742 Free PMC article.
-
Mechanism and components of endoplasmic reticulum-associated degradation.J Biochem. 2010 Jan;147(1):19-25. doi: 10.1093/jb/mvp194. Epub 2009 Nov 18. J Biochem. 2010. PMID: 19923195 Review.
-
Protein Misfolding and Aggregation: The Relatedness between Parkinson's Disease and Hepatic Endoplasmic Reticulum Storage Disorders.Int J Mol Sci. 2021 Nov 18;22(22):12467. doi: 10.3390/ijms222212467. Int J Mol Sci. 2021. PMID: 34830348 Free PMC article. Review.
Cited by
-
Autophagy and ethanol-induced liver injury.World J Gastroenterol. 2009 Mar 14;15(10):1178-85. doi: 10.3748/wjg.15.1178. World J Gastroenterol. 2009. PMID: 19291817 Free PMC article. Review.
-
Novel variant fibrinogen γp.C352R produced hypodysfibrinogenemia leading to a bleeding episode and failure of infertility treatment.Int J Hematol. 2021 Sep;114(3):325-333. doi: 10.1007/s12185-021-03174-y. Epub 2021 Jun 12. Int J Hematol. 2021. PMID: 34117991
-
Endoplasmic reticulum stress regulation of the Kar2p/BiP chaperone alleviates proteotoxicity via dual degradation pathways.Mol Biol Cell. 2012 Feb;23(4):630-41. doi: 10.1091/mbc.E11-04-0297. Epub 2011 Dec 21. Mol Biol Cell. 2012. PMID: 22190740 Free PMC article.
-
Structural Characteristics in the γ Chain Variants Associated with Fibrinogen Storage Disease Suggest the Underlying Pathogenic Mechanism.Int J Mol Sci. 2020 Jul 20;21(14):5139. doi: 10.3390/ijms21145139. Int J Mol Sci. 2020. PMID: 32698516 Free PMC article.
-
Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability.Am J Pathol. 2007 Aug;171(2):513-24. doi: 10.2353/ajpath.2007.070188. Epub 2007 Jul 9. Am J Pathol. 2007. PMID: 17620365 Free PMC article.
References
-
- Mosesson MW. Fibrinogen and fibrin structure and function. J Thromb Haemost. 2005;3:1894–1904. - PubMed
-
- Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. - PubMed
-
- Roy S, Yu S, Banerjee D, Overton O, Mukhopadhyay G, Oddoux C, Grieninger G, Redman CM. Assembly and secretion of fibrinogen: degradation of individual chains. J Biol Chem. 1992;267:23151–23158. - PubMed
-
- Huang S, Cao Z, Chung DW, Davie EW. The role of betagamma and alphagamma complexes in the assembly of human fibrinogen. J Biol Chem. 1996;271:27942–27947. - PubMed
-
- Redman CM, Xia H. Fibrinogen biosynthesis: assembly, intracellular degradation, and association with lipid synthesis and secretion. Ann NY Acad Sci. 2001;936:480–495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

