Proteomic profiling of metalloprotease activities with cocktails of active-site probes
- PMID: 16565715
- PMCID: PMC1538544
- DOI: 10.1038/nchembio781
Proteomic profiling of metalloprotease activities with cocktails of active-site probes
Abstract
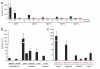
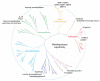
Metalloproteases are a large, diverse class of enzymes involved in many physiological and disease processes. Metalloproteases are regulated by post-translational mechanisms that diminish the effectiveness of conventional genomic and proteomic methods for their functional characterization. Chemical probes directed at active sites offer a potential way to measure metalloprotease activities in biological systems; however, large variations in structure limit the scope of any single small-molecule probe aimed at profiling this enzyme class. Here, we address this problem by creating a library of metalloprotease-directed probes that show complementary target selectivity. These probes were applied as a 'cocktail' to proteomes and their labeling profiles were analyzed collectively using an advanced liquid chromatography-mass spectrometry platform. More than 20 metalloproteases were identified, including members from nearly all of the major branches of this enzyme class. These findings suggest that chemical proteomic methods can serve as a universal strategy to profile the activity of the metalloprotease superfamily in complex biological systems.
Figures



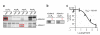
Comment in
-
Metalloproteases see the light.Nat Chem Biol. 2006 May;2(5):229-30. doi: 10.1038/nchembio0506-229. Nat Chem Biol. 2006. PMID: 16619018 No abstract available.
Similar articles
-
Developing photoactive affinity probes for proteomic profiling: hydroxamate-based probes for metalloproteases.J Am Chem Soc. 2004 Nov 10;126(44):14435-46. doi: 10.1021/ja047044i. J Am Chem Soc. 2004. PMID: 15521763
-
Trifunctional chemical probes for the consolidated detection and identification of enzyme activities from complex proteomes.Mol Cell Proteomics. 2002 Oct;1(10):828-35. doi: 10.1074/mcp.t200007-mcp200. Mol Cell Proteomics. 2002. PMID: 12438565
-
Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype.Nat Biotechnol. 2002 Aug;20(8):805-9. doi: 10.1038/nbt714. Epub 2002 Jul 1. Nat Biotechnol. 2002. PMID: 12091914
-
Tagging and detection strategies for activity-based proteomics.Curr Opin Chem Biol. 2007 Feb;11(1):20-8. doi: 10.1016/j.cbpa.2006.11.030. Epub 2006 Dec 13. Curr Opin Chem Biol. 2007. PMID: 17174138 Review.
-
Probes for activity-based profiling of plant proteases.Physiol Plant. 2012 May;145(1):18-27. doi: 10.1111/j.1399-3054.2011.01528.x. Epub 2011 Nov 15. Physiol Plant. 2012. PMID: 21985675 Review.
Cited by
-
The Tumor Proteolytic Landscape: A Challenging Frontier in Cancer Diagnosis and Therapy.Int J Mol Sci. 2021 Mar 3;22(5):2514. doi: 10.3390/ijms22052514. Int J Mol Sci. 2021. PMID: 33802262 Free PMC article. Review.
-
A New Affinity-Based Probe to Profile MMP Active Forms.Methods Mol Biol. 2024;2747:29-39. doi: 10.1007/978-1-0716-3589-6_3. Methods Mol Biol. 2024. PMID: 38038929
-
Small-molecule probes elucidate global enzyme activity in a proteomic context.BMB Rep. 2014 Mar;47(3):149-57. doi: 10.5483/bmbrep.2014.47.3.264. BMB Rep. 2014. PMID: 24499666 Free PMC article. Review.
-
Effect of sulindac sulfide on metallohydrolases in the human colon cancer cell line HT-29.PLoS One. 2011;6(10):e25725. doi: 10.1371/journal.pone.0025725. Epub 2011 Oct 3. PLoS One. 2011. PMID: 21991341 Free PMC article.
-
Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation.J Am Chem Soc. 2014 Jun 18;136(24):8669-76. doi: 10.1021/ja502372j. Epub 2014 May 30. J Am Chem Soc. 2014. PMID: 24836640 Free PMC article.
References
-
- Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 2001;70:209–246. - PubMed
-
- Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nat. Chem. Biol. 2005;1:130–142. - PubMed
-
- Kobe B, Kemp BE. Active site-directed protein regulation. Nature. 1999;402:373–376. - PubMed
-
- Jessani N, Cravatt BF. The development and application of methods for activity-based protein profiling. Curr. Opin. Chem. Biol. 2004;8:54–59. - PubMed
Publication types
MeSH terms
Substances
Associated data
- PubChem-Substance/10318689
- PubChem-Substance/10318690
- PubChem-Substance/10318691
- PubChem-Substance/10318692
- PubChem-Substance/10318693
- PubChem-Substance/10318694
- PubChem-Substance/10318695
- PubChem-Substance/10318696
- PubChem-Substance/10318697
- PubChem-Substance/10318698
- PubChem-Substance/10318699
- PubChem-Substance/10318700
- PubChem-Substance/10318701
- PubChem-Substance/10318702
- PubChem-Substance/10318703
- PubChem-Substance/10318704
- PubChem-Substance/10318705
- PubChem-Substance/10318706
- PubChem-Substance/10318707
- PubChem-Substance/10318708
- PubChem-Substance/10318709
- PubChem-Substance/10318710
- PubChem-Substance/10318711
- PubChem-Substance/10318712
- PubChem-Substance/10318713
- PubChem-Substance/10318714
- PubChem-Substance/10318715
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

