Exploring predictive and reproducible modeling with the single-subject FIAC dataset
- PMID: 16565951
- PMCID: PMC6871419
- DOI: 10.1002/hbm.20243
Exploring predictive and reproducible modeling with the single-subject FIAC dataset
Abstract
Predictive modeling of functional magnetic resonance imaging (fMRI) has the potential to expand the amount of information extracted and to enhance our understanding of brain systems by predicting brain states, rather than emphasizing the standard spatial mapping. Based on the block datasets of Functional Imaging Analysis Contest (FIAC) Subject 3, we demonstrate the potential and pitfalls of predictive modeling in fMRI analysis by investigating the performance of five models (linear discriminant analysis, logistic regression, linear support vector machine, Gaussian naive Bayes, and a variant) as a function of preprocessing steps and feature selection methods. We found that: (1) independent of the model, temporal detrending and feature selection assisted in building a more accurate predictive model; (2) the linear support vector machine and logistic regression often performed better than either of the Gaussian naive Bayes models in terms of the optimal prediction accuracy; and (3) the optimal prediction accuracy obtained in a feature space using principal components was typically lower than that obtained in a voxel space, given the same model and same preprocessing. We show that due to the existence of artifacts from different sources, high prediction accuracy alone does not guarantee that a classifier is learning a pattern of brain activity that might be usefully visualized, although cross-validation methods do provide fairly unbiased estimates of true prediction accuracy. The trade-off between the prediction accuracy and the reproducibility of the spatial pattern should be carefully considered in predictive modeling of fMRI. We suggest that unless the experimental goal is brain-state classification of new scans on well-defined spatial features, prediction alone should not be used as an optimization procedure in fMRI data analysis.
Figures


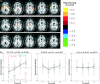

References
-
- Formisano E, Esposito F, Kriegeskorte N, Tedeschi G, Di Salle F, Goebel R (2002): Spatial independent component analysis of functional magnetic resonance imaging time‐series: characterization of the cortical components. Neurocomputing 49: 241–254.
-
- Gavrilescu M, Shaw ME, Stuart GW, Eckersley P, Svalbe ID, Egan GF (2002): Simulation of the effects of global normalization procedures in functional MRI. Neuroimage 17: 532–542. - PubMed
-
- Hastie T, Tibshirani R, Friedman J (2001): The elements of statistical learning theory. New York: Springer.
-
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P (2001): Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293: 2425–2430. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

