Identification and characterization of human polyserase-3, a novel protein with tandem serine-protease domains in the same polypeptide chain
- PMID: 16566820
- PMCID: PMC1435904
- DOI: 10.1186/1471-2091-7-9
Identification and characterization of human polyserase-3, a novel protein with tandem serine-protease domains in the same polypeptide chain
Abstract
Background: We have previously described the identification and characterization of polyserase-1 and polyserase-2, two human serine proteases containing three different catalytic domains within the same polypeptide chain. Polyserase-1 shows a complex organization and it is synthesized as a membrane-bound protein which can generate three independent serine protease domains as a consequence of post-translational processing events. The two first domains are enzymatically active. By contrast, polyserase-2 is an extracellular glycosylated protein whose three protease domains remain embedded in the same chain, and only the first domain possesses catalytic activity.
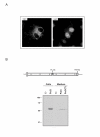
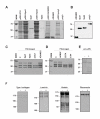
Results: Following our interest in the study of the human degradome, we have cloned a human liver cDNA encoding polyserase-3, a new protease with tandem serine protease domains in the same polypeptide chain. Comparative analysis of polyserase-3 with the two human polyserases described to date, revealed that this novel polyprotein is more closely related to polyserase-2 than to polyserase-1. Thus, polyserase-3 is a secreted protein such as polyserase-2, but lacks additional domains like the type II transmembrane motif and the low-density lipoprotein receptor module present in the membrane-anchored polyserase-1. Moreover, analysis of post-translational mechanisms operating in polyserase-3 maturation showed that its two protease domains remain as integral parts of the same polypeptide chain. This situation is similar to that observed in polyserase-2, but distinct from polyserase-1 whose protease domains are proteolytically released from the original chain to generate independent units. Immunolocalization studies indicated that polyserase-3 is secreted as a non-glycosylated protein, thus being also distinct from polyserase-2, which is a heavily glycosylated protein. Enzymatic assays indicated that recombinant polyserase-3 degrades the alpha-chain of fibrinogen as well as pro-urokinase-type plasminogen activator (pro-uPA). Northern blot analysis showed that polyserase-3 exhibits a unique expression pattern among human polyserases, being predominantly detected in testis, liver, heart and ovary, as well as in several tumor cell lines.
Conclusion: These findings contribute to define the growing group of human polyserine proteases composed at present by three different proteins. All of them share a complex structural design with several catalytic units in a single polypeptide but also show specific features in terms of enzymatic properties, expression patterns and post-translational maturation mechanisms.
Figures




Similar articles
-
Human polyserase-2, a novel enzyme with three tandem serine protease domains in a single polypeptide chain.J Biol Chem. 2005 Jan 21;280(3):1953-61. doi: 10.1074/jbc.M409139200. Epub 2004 Nov 9. J Biol Chem. 2005. PMID: 15536082
-
Polyserase-I, a human polyprotease with the ability to generate independent serine protease domains from a single translation product.Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9185-90. doi: 10.1073/pnas.1633392100. Epub 2003 Jul 28. Proc Natl Acad Sci U S A. 2003. PMID: 12886014 Free PMC article.
-
Expanding the complexity of the human degradome: polyserases and their tandem serine protease domains.Front Biosci. 2007 May 1;12:4661-9. doi: 10.2741/2415. Front Biosci. 2007. PMID: 17485402 Review.
-
Serase-1B, a new splice variant of polyserase-1/TMPRSS9, activates urokinase-type plasminogen activator and the proteolytic activation is negatively regulated by glycosaminoglycans.Biochem J. 2006 Dec 15;400(3):551-61. doi: 10.1042/BJ20060212. Biochem J. 2006. PMID: 16872279 Free PMC article.
-
Roles and regulation of membrane-associated serine proteases.Biochem Soc Trans. 2007 Jun;35(Pt 3):583-7. doi: 10.1042/BST0350583. Biochem Soc Trans. 2007. PMID: 17511657 Review.
Cited by
-
Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains.PLoS One. 2017 Nov 13;12(11):e0187778. doi: 10.1371/journal.pone.0187778. eCollection 2017. PLoS One. 2017. PMID: 29131831 Free PMC article.
-
Oral anticoagulation and VKORC1 polymorphism in patients with a mechanical heart prosthesis: a 6-year follow-up.J Thromb Thrombolysis. 2012 Nov;34(4):506-12. doi: 10.1007/s11239-012-0740-8. J Thromb Thrombolysis. 2012. PMID: 22592842 Clinical Trial.
-
Identification of protease serine S1 family member 53 as a mitochondrial protein in murine islet beta cells.Islets. 2022 Jan 1;14(1):1-13. doi: 10.1080/19382014.2021.1982325. Epub 2021 Oct 12. Islets. 2022. PMID: 34636707 Free PMC article.
-
Genome-wide association analysis identifies three psoriasis susceptibility loci.Nat Genet. 2010 Nov;42(11):1000-4. doi: 10.1038/ng.693. Epub 2010 Oct 17. Nat Genet. 2010. PMID: 20953189 Free PMC article.
-
Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity.Nat Genet. 2012 Dec;44(12):1341-8. doi: 10.1038/ng.2467. Epub 2012 Nov 11. Nat Genet. 2012. PMID: 23143594 Free PMC article.
References
-
- Eberhardt RY, Gilbert HJ, Hazlewood GP. Primary sequence and enzymic properties of two modular endoglucanases, Cel5A and Cel45A, from the anaerobic fungus Piromyces equi. Microbiology. 2000;146 ( Pt 8):1999–2008. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

