Simulation of polymer translocation through protein channels
- PMID: 16567657
- PMCID: PMC1551897
- DOI: 10.1073/pnas.0510725103
Simulation of polymer translocation through protein channels
Abstract
A modeling algorithm is presented to compute simultaneously polymer conformations and ionic current, as single polymer molecules undergo translocation through protein channels. The method is based on a combination of Langevin dynamics for coarse-grained models of polymers and the Poisson-Nernst-Planck formalism for ionic current. For the illustrative example of ssDNA passing through the alpha-hemolysin pore, vivid details of conformational fluctuations of the polymer inside the vestibule and beta-barrel compartments of the protein pore, and their consequent effects on the translocation time and extent of blocked ionic current are presented. In addition to yielding insights into several experimentally reported puzzles, our simulations offer experimental strategies to sequence polymers more efficiently.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

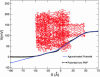


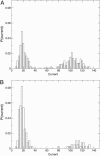

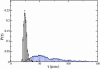

Similar articles
-
Langevin dynamics simulation on the translocation of polymer through α-hemolysin pore.J Phys Condens Matter. 2014 Oct 15;26(41):415101. doi: 10.1088/0953-8984/26/41/415101. Epub 2014 Sep 5. J Phys Condens Matter. 2014. PMID: 25192215
-
Simulations of stochastic sensing of proteins.J Am Chem Soc. 2005 Dec 28;127(51):18252-61. doi: 10.1021/ja055695o. J Am Chem Soc. 2005. PMID: 16366579
-
Role of non-equilibrium conformations on driven polymer translocation.J Chem Phys. 2018 Jan 14;148(2):024903. doi: 10.1063/1.4994204. J Chem Phys. 2018. PMID: 29331138 Free PMC article.
-
Theoretical Modeling of Polymer Translocation: From the Electrohydrodynamics of Short Polymers to the Fluctuating Long Polymers.Polymers (Basel). 2019 Jan 11;11(1):118. doi: 10.3390/polym11010118. Polymers (Basel). 2019. PMID: 30960102 Free PMC article. Review.
-
Single-polymer dynamics under constraints: scaling theory and computer experiment.J Phys Condens Matter. 2011 Mar 16;23(10):103101. doi: 10.1088/0953-8984/23/10/103101. Epub 2011 Feb 18. J Phys Condens Matter. 2011. PMID: 21335636 Review.
Cited by
-
Laser-based temperature control to study the roles of entropy and enthalpy in polymer-nanopore interactions.Sci Adv. 2021 Apr 21;7(17):eabf5462. doi: 10.1126/sciadv.abf5462. Print 2021 Apr. Sci Adv. 2021. PMID: 33883140 Free PMC article.
-
Directly observing the motion of DNA molecules near solid-state nanopores.ACS Nano. 2012 Nov 27;6(11):10090-7. doi: 10.1021/nn303816w. Epub 2012 Oct 12. ACS Nano. 2012. PMID: 23046052 Free PMC article.
-
Determination of Molecular Weights in Polyelectrolyte Mixtures Using Polymer Translocation through a Protein Nanopore.ACS Macro Lett. 2014 Sep 16;3(9):911-915. doi: 10.1021/mz500404e. Epub 2014 Sep 2. ACS Macro Lett. 2014. PMID: 25243100 Free PMC article.
-
Effects of Nanopore Charge Decorations on the Translocation Dynamics of DNA.Biophys J. 2017 Oct 17;113(8):1664-1672. doi: 10.1016/j.bpj.2017.08.045. Biophys J. 2017. PMID: 29045861 Free PMC article.
-
Enzyme-modulated DNA translocation through a nanopore.J Am Chem Soc. 2009 Dec 30;131(51):18563-70. doi: 10.1021/ja904047q. J Am Chem Soc. 2009. PMID: 19958025 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

