Simulation of polymer translocation through protein channels
- PMID: 16567657
- PMCID: PMC1551897
- DOI: 10.1073/pnas.0510725103
Simulation of polymer translocation through protein channels
Abstract
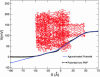
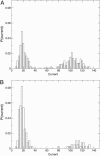
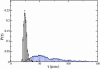
A modeling algorithm is presented to compute simultaneously polymer conformations and ionic current, as single polymer molecules undergo translocation through protein channels. The method is based on a combination of Langevin dynamics for coarse-grained models of polymers and the Poisson-Nernst-Planck formalism for ionic current. For the illustrative example of ssDNA passing through the alpha-hemolysin pore, vivid details of conformational fluctuations of the polymer inside the vestibule and beta-barrel compartments of the protein pore, and their consequent effects on the translocation time and extent of blocked ionic current are presented. In addition to yielding insights into several experimentally reported puzzles, our simulations offer experimental strategies to sequence polymers more efficiently.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

