Structure-activity relationships of aminocoumarin-type gyrase and topoisomerase IV inhibitors obtained by combinatorial biosynthesis
- PMID: 16569821
- PMCID: PMC1426943
- DOI: 10.1128/AAC.50.4.1136-1142.2006
Structure-activity relationships of aminocoumarin-type gyrase and topoisomerase IV inhibitors obtained by combinatorial biosynthesis
Abstract
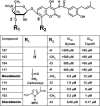
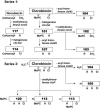
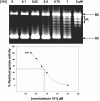
Novobiocin and clorobiocin are gyrase inhibitors produced by Streptomyces strains. Structurally, the two compounds differ only by substitution at two positions: CH3 versus Cl at position 8' of the aminocoumarin ring and carbamoyl versus 5-methyl-pyrrol-2-carbonyl (MePC) at the 3"-OH of noviose. Using genetic engineering, we generated a series of analogs carrying H, CH3, or Cl at 8' and H, carbamoyl, or MePC at 3"-OH. Comparison of the gyrase inhibitory activities of all nine structural permutations confirmed that acylation of 3"-OH is essential for activity, with MePC being more effective than carbamoyl. Substitution at 8' further enhanced activity, but the effect of CH3 or Cl depended on the nature of the acyl group at 3": in the presence of carbamoyl at 3", CH3 resulted in higher activity; in the presence of MePC at 3", Cl resulted in higher activity. This suggests that the structures of both natural compounds are highly evolved for optimal interaction with gyrase. In a second series of experiments, clorobiocin derivatives with and without the methyl group at 4"-OH of noviose, and with different positions of the MePC group of noviose, were tested. Again clorobiocin was superior to all of its analogs. The activities of all compounds were also tested against topoisomerase IV (topo IV). Clorobiocin stood out as a remarkably effective topo IV inhibitor. The relative activities of the different compounds toward topo IV showed a pattern similar to that of the relative gyrase-inhibitory activities. This is the first report of a systematic evaluation of a series of aminocoumarins against both gyrase and topo IV. The results give further insight into the structure-activity relationships of aminocoumarin antibiotics.
Figures


Similar articles
-
Biological activities of novel gyrase inhibitors of the aminocoumarin class.Antimicrob Agents Chemother. 2008 Jun;52(6):1982-90. doi: 10.1128/AAC.01235-07. Epub 2008 Mar 17. Antimicrob Agents Chemother. 2008. PMID: 18347114 Free PMC article.
-
Inhibition of DNA gyrase and DNA topoisomerase IV of Staphylococcus aureus and Escherichia coli by aminocoumarin antibiotics.J Antimicrob Chemother. 2011 Sep;66(9):2061-9. doi: 10.1093/jac/dkr247. Epub 2011 Jun 21. J Antimicrob Chemother. 2011. PMID: 21693461
-
New aminocoumarin antibiotics as gyrase inhibitors.Int J Med Microbiol. 2014 Jan;304(1):31-6. doi: 10.1016/j.ijmm.2013.08.013. Epub 2013 Sep 4. Int J Med Microbiol. 2014. PMID: 24079980 Review.
-
Effects of novobiocin, coumermycin A1, clorobiocin, and their analogs on Escherichia coli DNA gyrase and bacterial growth.Antimicrob Agents Chemother. 1982 Oct;22(4):662-71. doi: 10.1128/AAC.22.4.662. Antimicrob Agents Chemother. 1982. PMID: 6295263 Free PMC article.
-
New aminocoumarin antibiotics from genetically engineered Streptomyces strains.Curr Med Chem. 2005;12(4):419-27. doi: 10.2174/0929867053363063. Curr Med Chem. 2005. PMID: 15720250 Review.
Cited by
-
Identification of ATP-Competitive Human CMG Helicase Inhibitors for Cancer Intervention that Disrupt CMG-Replisome Function.Mol Cancer Ther. 2024 Nov 4;23(11):1568-1585. doi: 10.1158/1535-7163.MCT-23-0904. Mol Cancer Ther. 2024. PMID: 38982858 Free PMC article.
-
Biological activities of novel gyrase inhibitors of the aminocoumarin class.Antimicrob Agents Chemother. 2008 Jun;52(6):1982-90. doi: 10.1128/AAC.01235-07. Epub 2008 Mar 17. Antimicrob Agents Chemother. 2008. PMID: 18347114 Free PMC article.
-
Towards Conformation-Sensitive Inhibition of Gyrase: Implications of Mechanistic Insight for the Identification and Improvement of Inhibitors.Molecules. 2021 Feb 25;26(5):1234. doi: 10.3390/molecules26051234. Molecules. 2021. PMID: 33669078 Free PMC article. Review.
-
The crystal structure of the novobiocin biosynthetic enzyme NovP: the first representative structure for the TylF O-methyltransferase superfamily.J Mol Biol. 2010 Jan 15;395(2):390-407. doi: 10.1016/j.jmb.2009.10.045. Epub 2009 Oct 24. J Mol Biol. 2010. PMID: 19857499 Free PMC article.
-
Exploiting bacterial DNA gyrase as a drug target: current state and perspectives.Appl Microbiol Biotechnol. 2011 Nov;92(3):479-97. doi: 10.1007/s00253-011-3557-z. Epub 2011 Sep 9. Appl Microbiol Biotechnol. 2011. PMID: 21904817 Free PMC article. Review.
References
-
- Drlica, K., and M. Malik. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:249-282. - PubMed
-
- Eustaquio, A. S., B. Gust, S. M. Li, S. Pelzer, W. Wohlleben, K. F. Chater, and L. Heide. 2004. Production of 8′-halogenated and 8′-unsubstituted novobiocin derivatives in genetically engineered Streptomyces coelicolor strains. Chem. Biol. 11:1561-1572. - PubMed
-
- Eustaquio, A. S., B. Gust, T. Luft, S. M. Li, K. F. Chater, and L. Heide. 2003. Clorobiocin biosynthesis in Streptomyces. Identification of the halogenase and generation of structural analogs. Chem. Biol. 10:279-288. - PubMed
-
- Freitag, A., H. Rapp, L. Heide, and S. M. Li. 2005. Metabolic engineering of aminocoumarins: inactivation of the methyltransferase gene cloP and generation of new clorobiocin derivatives in a heterologous host. Chembiochem 6:1411-1418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

